The role of microRNAs in prostate cancer progression
Introduction
Prostate cancer (PCa) is a typical hormone-dependent disease (1); however, almost all PCa patients with androgen-ablation therapy ultimately become castration-resistant prostate cancer (CRPC), which contributes to the majority of mortality in PCa. Androgen receptor (AR) is known as a key mitogen for the growth and morphogen for the development of prostate. In PCa, AR activity is critical for the disease progression and becomes hyperactivated in CRPC. Mounting evidences indicate that the development of PCa is highly associated with aberrant AR activity resulted from AR gene amplification and/or mutation, alternative splicing (2-7), the cross talk with growth factor signaling pathways (8-13), and the presence of AR co-activators and AR co-repressors (14-17). In particular, Dicer-mediated maturation of microRNAs (miRNAs) suppresses the expression level of AR co-repressors, NCoR and SMRT, leading to enhance AR transcriptional activity (18). Based on the interaction between Dicer and AR, the correlation between AR and microRNA signaling has been broadly examined to investigate the fundamental role of miRNAs in PCa progression.
miRNAs are a large family of small 20-25 nt single stranded noncoding RNAs, which can interfere with the expression of ~60% protein coding genes by post-transcriptional suppression, target mRNA degradation, or translational inhibition (19). In the past two decades, significant advances have been achieved in miRNA research. miRNAs are found to be highly conserved among the animal phylogeny. Based on their conserved sequences, miRNAs shared an identical seed region of 2-7 nucleotides are grouped into different family. Up to date, 63 miRNA families have been categorized and more than 1,000 miRNAs have been fully characterized in their expression, epigenetic regulation, biogenesis and functions. In general, miRNAs are either derived from non-coding RNA transcripts or located within the introns of protein-coding genes (20,21). Multiple miRNAs can be clustered in close proximity and encoded together as a single polycistronic primary transcript, such as miR-106a-363 (22) and miR-17-92 clusters (23). The transcriptional mechanism of microRNA is similar to that of mRNA. miRNA gene promoters are regulated by transcriptional factors that also regulate protein-coding gene expression. For example, the promoter region of miR-21 can be regulated by AR, activation protein 1 (AP-1) and signal transducer and activator of transcription 3 (STAT-3) (24,25). Hence elevation of miR-21 in cancer is partially due to aberrant activation of AR and AP-1. Meanwhile, c-Myc is a well-known oncogene that is suggested to regulate an oncogenic miR-17-92 cluster. Overexpression of both c-Myc and miR-17-92 cluster is indicated to enhance the tumor aggressiveness (26). Moreover, the genomic organization of miRNAs reveals that about 52% of miRNA genes are localized at the fragile chromosomal regions, which are susceptible to amplification, deletion and translocation associated with cancer. A recent study indicates that let-7 miRNAs family is located in the genomic regions that are frequently deleted in multiple cancer types including PCa (27). Moreover, the miR-15a/miR-16-1 cluster is located at chromosome 13q14. The frequency of allelic loss at 13q increases from early, advanced to metastatic PCa (28). In addition, aberrant DNA hypermethylation at the CpG island is often observed in the promoter region or transcriptional start site of tumor suppressive miRNAs such as miR-200/-141, miR-205, miR-34, miR-143 and miR-145 associated with PCa (29-32). Thus epigenetic regulation is also a key regulatory mechanism for miRNA gene expression.
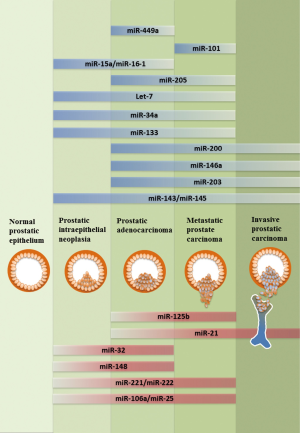
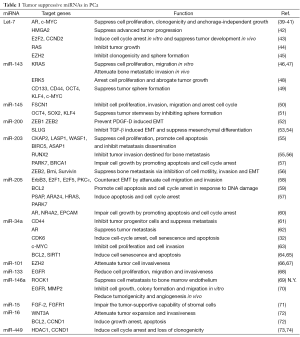
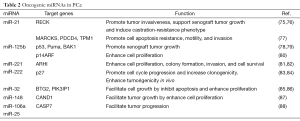
In addition to the regulation of miRNA gene expression, the biosynthetic process of miRNA maturation becomes an emerging area. The biogenesis of miRNAs composes sequential steps of RNase III-mediated endonucleolytic cleavage mechanisms (33,34). In brief, the primary transcripts of miRNAs are transcribed by RNA polymerase II and processed in the nucleus by Drosha and Pasha (DGCR8) into a 70-100 nucleotides-long precursor miRNAs (pre-miRNAs) (35). Pre-miRNAs are exported to the cytoplasm through Exportin 5 and further processed by Dicer, generating a 20-25 nt RNA duplex comprise of a matured guide strand and a complementary passenger strand (miRNA*). The single stranded matured miRNA is then incorporated into the RNA-induced silencing complex (RISC) associated with Agonaut (AGO2), and bound to the complementary sequence on the 3' untranslated region (3' UTR) of target mRNA, leading to mRNA degradation (36-38). Based on their post-transcriptional regulation on a variety of target genes, miRNA is expected to be involved in virtually every biologic process in cell. In cancer, based on their post-transcriptional repression on a variety of oncogenes or tumor suppressor genes, miRNAs are also divided into onco-miRNAs (oncomirs) and tumor suppressors miRNAs (Figure 1). Overall, the importance of microRNAs has become a key to gain more understanding of molecular mechanisms associated with prostatic carcinogenesis. In this review, we will focus on key unique miRNAs (Tables 1,2) involved in PCa.
Full table
Full table
Tumor suppressive miRNAs
Let-7 family
The let-7 gene encodes a highly conserved miRNA family of let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i and miR-98, which are significantly down-regulated in localized PCa, compared to adjacent benign tissues (39). The functionality of let-7 has been shown to target oncogenes involved in cell-cycle regulation, cell migration, proliferation, differentiation, and epithelial-to-mesenchymal transition (EMT) progression. In particular, let-7g can inhibit tumor growth via post-transcriptional suppression on RAS oncogene (44). On the other hand, loss of let-7 miRNAs is corresponded with elevated level of Enhancer of Zeste homolog 2 (Ezh2) correlated with PCa progression (45). Ectopic expression of let-7 results in the reduction of Ezh2, accompanied with diminished clonogenic ability and sphere formation in PCa cells (45). Another let-7 target gene is High-mobility group AT-hook 2 (HMGA2) (89) that is highly expressed in PCa compared to adjacent benign tissues. Indeed, HMGA2 was found de-repressed upon let-7 inhibition (43). Meanwhile, co-regulation of HMGA2 and Smad were found to orchestrate an EMT transcriptional network via targeting the promoter of SNAI1 in human hepatocarcinoma cell line (90). These results suggest a possibility that let-7 could inhibit EMT via targeting HMGA2 during PCa progression. Moreover, another study also imply that let-7 can induce cell cycle arrest and xenograft PCa tumor development by suppressing E2F2 and CCND2, which are found to be the direct target of let-7 (43). Lin-28 is a well-identified post-transcriptional suppressor of precursor let-7 maturation (91,92); An inverse correlation between lin28 and let-7 is also found in many cancer cell lines including PC3 (93). Based on these observations, lin28-mediated let-7 biogenesis has become an important mechanism to impact tumorigenesis. Conversely, let-7 can target the lin28 mRNA, suggesting that a reciprocal feedback loop exists between let-7 and lin28 (94-97). In addition, c-Myc is found to be a key factor involved in this interaction. c-Myc acts as a transcriptional activator for lin-28 gene expression and c-Myc is also found to be a target gene of let-7 family in multiple cancer types (40,98,99). Overall, the orchestrated interaction between lin28, let-7 and c-Myc is a complicated network of gene regulation, which is often altered in cancer cells (100). Also, let-7c is shown to antagonize AR expression by targeting c-Myc (101). Overexpression of let-7 leads to AR suppression, accompanied with attenuated cell proliferation, clonogenicity and anchorage-independent growth in PCa cells (39,41). Overall, the let-7 miRNA family exerts tumor suppressor characteristics via targeting multiple oncogenes including RAS, HMGA2, Ezh2, Lin28 and c-Myc. Therefore, let-7 could be a potential diagnostic biomarker and further developed into a new therapeutic strategy for PCa.
miR-143 and miR-145
Both miR-143 and miR-145 are derived from the same miR-143/-145 cluster, which are found down-regulated in metastatic PCa samples (29). Both miR-143 and miR-145 share similar functions in tumor suppression. First, miR-143 is found to exhibit a negative effect on PCa cell proliferation and migration by targeting ERK5 and KRAS, and inactivating subsequent epidermal growth factor receptor (EGFR)-RAS-MAPK signaling pathway (46,48). On the other hand, miR-145 is shown to inhibit PCa cell proliferation by targeting Fascin homolog 1 (FSCN1) that is an actin bundling protein involved in cell motility, adhesion and cellular interactions during tumorigenesis and metastasis (50). Second, overexpression of both miRNAs in PC3 cells represses fibronectin and enhances E-cadherin expression and both can reverse EMT and further attenuate the tumor invasiveness in an in vivo bone metastasis model (47). Third, a recent study indicates that both miR-143 and miR-145 can suppress the stem cell characteristics in PC3 cell lines by inhibiting the stem cell markers or factors including CD133, CD44, Oct4, c-Myc and Klf4 (49). Similarly, some studies of embryonic stem cell (ESCs) indicate that miR-145 has been identified to repress pluripotency by targeting Oct4, Sox2, and Klf4 (51,102). Taken together, both miR-143 and miR-145 can suppress several cancer behaviors of PCa cells from tumor proliferation, invasion/metastasis and stemness.
miR-200 family
During embryogenesis, EMT is established to determine the transition between epithelial and mesenchymal phenotypes at different developmental stages (103,104). However, during prostatic carcinogenesis, EMT has been highly implicated in PCa progression by initiating the tumor invasiveness (105-107). The consequences of EMT result in the suppression of epithelial markers by transcriptional repressors including ZEB1, ZEB2, SNAI1 and SNAI2, which are found to be the target genes of several tumor suppressive miRNAs including the miR-200 family. The miR-200 family consists of miR-200a, miR-200b, miR-200c, miR-141 and miR-449, which are significantly down-regulated during PCa progression and identified to suppress PCa tumor metastasis particularly via inhibiting EMT. A recent study using PC3 cell line indicates that miR-200 can inhibit the platelet-derived growth factor-D (PDGF-D)-induced acquisition of EMT via targeting of both ZEB1 and ZEB2 (52). Another group studying benign prostate hyperplasia (BPH) also shows that miR-200 can reverse the TGFβ-induced EMT phenotype in BPH cell line (53). Meanwhile, in kidney epithelial cell line, all miR-200 family members have been shown to suppress TGFβ-induced EMT via targeting ZEB1, ZEB2 and SNAI2 in vitro (108,109); Similar result is also found in unilateral urethral obstruction (UUO) model that miR-200 can protect renal tubular epithelial cells from mesenchymal transition via suppressing ZEB1 and ZEB2 in vivo (110). In addition, a regulatory feedback loop has been demonstrated between SNAI2 and the miR-200 family. While miR-200 can target SNAI2 mRNA, SNAI2 protein acts as a repressor to suppress miR-200 gene expression (54). Thus, down-regulation of miR-200 may disrupt the homeostasis between SNAI2 and miR-200. Overall, loss of the miR-200 family in PCa initiates EMT process, which is critical for PCa invasiveness.
miR-203
miR-203 is a well-characterized tumor suppressor and shared the similar anti-metastatic function to miR-200 family (111,112). MiR-203 has been demonstrated to induce MET in PC3 and DU145 cell lines via targeting CKAP2, LASP1, BIRC5, WASF1, ASAP1, and RUNX2, which are critical effectors involved in cell proliferation, migration, invasion and EMT (55). Meanwhile, other study also suggests that miR-203 exhibits its negative effect on multiple steps of the PCa metastatic cascade via targeting on pro-metastatic molecules including ZEB2, Bmi, survivin, and Runx2. As a result, restoration of miR-203 in PC3, VCaP, and MDA-PCa-2b cell lines attenuates the invasiveness of PCa bone metastasis in vivo (56). This evidence suggests miR-203 play an important role in the metastatic progression of PCa and that loss of miR-203 may further enhance the invasive characteristics of advanced PCa.
miR-205
Similar to miR-203, miR-205 regulates PCa progression by targeting EMT signaling mechanisms (113). Restoration of miR-205 in PCa cells can induce MET phenotype by up-regulation of E-cadherin, along with attenuated cell invasiveness. In a more detailed study, miR-205 is suggested to attenuate cell invasion and migration via targeting ErbB3, E2F1, E2F5 and protein kinase Cε (PKCε) (58). Meanwhile, another study using xenograft model with tail vein injection also demonstrates that miR-205 can inhibit PCa lung metastasis in vivo by targeting ZEB1 and vimentin (114). In addition to EMT regulation, miR-205 can also promote PCa cell apoptosis and cell-cycle arrest by targeting the anti-apoptotic gene BCL2 (59). Moreover, miR-205 is able to inhibit tumor cell growth by inducing apoptosis and cell cycle arrest via targeting AR co-regulators (DJ-1, PSAP, ARA24) and MAPK signaling components (57). These accumulating findings indicate that both miR-203 and miR-205 may suppress metastatic progression of PCa by impairing the EMT-induced invasiveness.
miR-34a
In PCa, miR-34a is identified as a tumor suppressor by inhibiting the stemness characteristics of prostate cancer stem cells (CSC) (61). The study demonstrates that miR-34a is down-regulated in the CD44 + PCa cells purified from xenograft tumors; overexpression of miR-34a can attenuate clonogenic expansion, tumor regeneration, and metastasis in CD44 + PCa cells. These results suggest that miR-34a is a negative regulator of prostate CSC and may exert its suppressive effect on PCa progression before the onset of metastatic CRPC. Moreover, miR-34a appears to be a p53-regulated gene from a study (115) using doxorubicin and camptothecin-induced p53 activation that a significant up-regulation of both miR-34a and miR-34c is shown. However, this p53-mediated miR-34a up-regulation is abolished in both AR-knockdown LNCaP cells and AR-negative cell lines including PC3 and DU145, suggesting miR-34a expression is AR-dependent. On the other hand, AR has been identified as a direct target gene of miR-34a (62); AR activity can be repressed by de-methylation of epigenetically silenced miR-34a promoter in PCa cells. Overall, these findings imply a reciprocal transcriptional regulatory network among miR-34a, p53 and AR in PCa cells. However, whether this network is involved in PCa progression and its clinical significance require further investigation. Another target gene of miR-34a is c-Myc (63). By targeting the c-Myc expression, miR-34a is shown to suppress the signaling cascade of c-Myc-Skp2-Miz1, which leads to RhoA gene expression and subsequent attenuates cell migration and invasion. In addition to PCa stemness and metastasis, miR-34a also affect the PCa tumor growth by inducing cell-cycle arrest, cell senescence and apoptosis via targeting cell-cycle regulatory gene, such as CDK6 (32), and anti-apoptosis genes including Bcl-2 and SIRT1 (64,65). Overall, miR-34a may exert its tumor suppressor role via targeting various signaling molecules at different stages of PCa progression.
miR-101
Ezh2 is a histone methyltransferase that regulates epigenetic silencing and early studies have demonstrated that overexpression of Ezh2 in PCa contributes to the enhanced aggressiveness and metastatic potential of PCa cells (116-119). A study shows an inverse correlation between miR-101 and Ezh2 expression has been observed in human PCa. Meanwhile, miR-101 is found to suppress the expression and function of Ezh2 in PCa cell lines (66) and the overexpression of miR-101 in PC3, DU145 and LNCaP cells also results in the suppression of Ezh2, along with attenuated cell invasion and migration of these PCa cell lines in vitro (67,117). These findings clearly indicate the role of miR-101 in the epigenetic regulation critical for PCa progression via targeting Ezh2 expression.
miR-133 and miR-146a
Epidermal growth factor (EGF) and EGFR are known to be key tumor promoter for PCa (120). A recent study indicates that, under hypoxia condition, EGFR can interrupt the biogenesis of mHESM (miRNAs regulated by hypoxia-dependent EGFR-suppressed maturation) resulted in reducing Dicer binding and abolishing miRNA maturation via targeting AGO2 phosphorylation (121). Implying that EGFR may certainly contribute to the modification of miRNA processing. On the other hand, both miR-133 and miR-146a have been shown to suppress PCa tumor progression via targeting EGFR. Down-regulation of miR-133 has been observed in PC3 and DU-145 cell lines. Ectopic expression of miR-133 can reduce cell proliferation, migration and invasiveness by targeting EGFR (68). Similar to miR-133, expression level of miR-146a is also significantly down-regulated in PCa (122,123). Overexpression of miR-146a has been demonstrated to suppress PCa cell growth, colony formation and migration in vivo via targeting EGFR. Additional studies also reveal that miR-146a can inhibit angiogenesis and bone metastasis in vivo by suppressing both matrix metalloproteinase-2 (MMP2) and Rho-associated, coiled-coil containing protein kinase 1 (ROCK1) expression (69,70). These findings indicate that loss of both miR-133 and miR-146a in PCa may attribute to enhancement of EGFR signaling, leading to aggressive PCa progression.
miR-15a and miR-16-1
miR-15a and miR-16-1 are in the same cluster; the expression of miR-15a/miR-16-1 is often down-regulated in PCa due to chromosomal deletion at 13q14, which is highly correlated with the progression of PCa (124). A study has demonstrated that miR-15a/miR-16-1 level is inversely correlated with B-cell lymphoma 2 (BCL2), cyclin-D1 (CCND1) and wingless-type 3A (WNT3A) in advanced PCa (72). The same group also found that both CCND1 and WNT3A are putative target genes of miR-15a/miR-16-1. As a result, restoration of miR-15a and miR-16 is shown to arrest cell growth and induce apoptosis and knockdown of mir-15a/miR-16-1 can promote survival, proliferation and invasiveness of PCa xenograft tumor in vivo. On the other hand, miR-15a and miR-16-1 also exert tumor suppressive effects by interfering the stromal support in the tumor microenvironment since interaction between tumor cells and the surrounding cellular components is critical for tumor development (125,126). It has been indicated that down-regulation of miR-15a and miR-16 in cancer-associated fibroblasts results in tumor expansion and invasion (72), which is supported by the reconstitution of both miRNAs in fibroblast can interrupt the stromal support by targeting FGF-2 and FGFR1 (71). All these data indicate that the tumor suppressor role of miR-15a/ miR-16-1 is to suppress cancer cells or interrupt their communication with the microenvironment.
miR-449a
miR-449a is inversely correlated with the expression of histone deacetylase 1 (HDAC1)-an enzyme critical for epigenetic regulation. It has been indicated that increased expression of miR-449a in PCa cell lines leads to both cell cycle arrest and loss of clonogenicity by targeting HDAC1 (73). In addition, miR-449 can initiate cell cycle arrest and induce cell senescence by targeting cyclinD1 (74).
Oncogenic miRNAs
miR-21
The recurrence of CRPC is often associated with hyperactivation of AR. Recent studies have suggested that several oncogenic miRNAs is correlated with aberrant AR activation. In particular, miR-21 is an AR-regulated miRNA and its expression level is consistently elevated from androgen-dependent PCa to CRPC (127). Overexpression of miR-21 can support xenograft tumor growth and induce castration-resistant phenotype (75). In addition to androgen response element (ARE), other cis-elements such as AP-1 and STAT-3 are also found in the promoter region of miR-21 (24,25). AP-1 activity is closely associated with CRPC recurrence (128) and STAT-3 is also shown to be involved in PCa metastasis (129). Overall, the highly elevation of miR-21 may be attributed to the aberrant expression of transcriptional activators such as AR and AP-1. The subsequent effect of miR-21 overexpression in turn contributes to the development of prostate tumorigenesis. Several target genes of miR-21 have been shown to suppress tumor progression by inhibiting invasiveness, promoting apoptosis and cell cycle arrest. For example, myristoylated alanine-rich protein kinase c substrate (MARCKS) is a direct target of miR-21, which plays key role in cell motility, membrane trafficking and mitogenesis. Thus, miR-21 promotes the apoptosis resistance, cell motility and invasiveness of PC3 and DU-145 cells partly via targeting MARCKS (77). Meanwhile, a recent study demonstrates that reversion-inducing cysteine-rich protein with Kazal motifs (RECK) is another novel target of miR-21; An inverse correlation between RECK and miR-21 has been shown from different stages of PCa (76).
miR-125b
Similar to miR-21, miR-125b is also an AR-induced miRNA. The induction of miR-125b in LNCaP cells inhibits apoptosis and enhances cell proliferation. Mechanistically, miR-125b promotes PCa xenograft tumor growth by targeting major pro-apoptotic genes including p53, Puma and BAK1 (78). Consistent with this observation, miR-125b is shown to modulate the p53 network by interrupting Mdm2 degradation via targeting p14ARF, which mediates the Mdm2 sequestration (80). Overall, miR-125b can target the p53-p21 and Puma signaling network, leading to enhanced cell proliferation in both LNCaP and CWR22Rv1 PCa cell lines through p53-dependent and p53-independent manner, respectively.
miR-221 and miR-222
Both miR-221 and miR-222 belong to the same miRNA cluster. Overexpression of miR-221/miR-222 has been often found in PCa. The aberrant elevation of miR-221/miR-222 is highly correlated with metastatic CRPC phenotypes. Moreover, an inverse correlation between miR-221/miR-222 expression and p27Kip1 level has been observed in primary PCa. Several studies demonstrated that miR-221/miR-222 can up-regulate S-phase kinase associated protein 2 (Skp2), cyclin A and cyclin D1via targeting p27Kip1 suppression, leading to cell cycle progression at G1-to -S phase, increased clonogenicity in vitro and enhanced tumorigenicity in vivo (83,84,130). Meanwhile, Ras homolog member I (ARHI), a tumor suppressor identified in ovarian cancer (131), is also identified as the target gene of miR-221/miR-222. Overexpression of ARHI in PC3 cells results in the inhibition of cell proliferation, colony formation, cell invasion and survival (81,82,132), suggesting decreased ARHI mRNA could be an additional mechanism for miR-221/miR-222 contributing to the accelerated tumor growth in PCa. Thus, these data conclude the functional role of miR-221/miR-222 as PCa promoter by targeting tumor suppressor genes such as p27Kip1 and ARHI.
miR-32
miR-32 is highly expressed in CRPC specimens compared to BPH specimens (85). A study demonstrated that miR-32 exerts oncogenic characteristics by targeting on both B-cell translocation gene 2 (BTG-2) and phosphoinositide-3-kinase interacting protein 1 (PIK3IP1), which regulates the inhibition of PI3K, a well-known regulator of cell proliferation, migration and survival (85). An inverse correlation between miR-32 and BTG-2 has been found in the CRPC specimens (85). In addition, numerous studies have identified BTG-2 as a critical target gene of AR-regulated miRNAs including miR-32, miR-148 and miR-21 (85,86). Loss of BTG-2 was implicated in the progression of PCa accompanied by the appearance of EMT markers (133). Meanwhile, a study using LNCaP cells demonstrated that miR-32 facilitates cell growth by inhibiting cell apoptosis and enhancing cell proliferation, respectively. Overall, miR-32 exerts its oncogenic characteristics by targeting on tumor suppressors critical for cell proliferation, survival and migration.
miR-148a
Similar to miR-32, miR-148 is elevated in advanced PCa compared to primary tumor (134). However, in contrast to the distinctive oncogenic role of miR-32, the role of miR-148a in PCa progression is more controversial. For example, miR-148a was identified as an androgen-responsive miRNA and facilitates LNCaP cell proliferation via targeting cullin-associated and neddylation-dissociated 1 (CAND1) (87). In contrast, miR-148a is shown to be down-regulated in both DU-145 and PC3 cell lines. Furthermore, overexpression of miR-148a in PC3 cells attenuates cell growth, migration, invasion, and enhances the drug sensitivity to Paclitaxel. This phenomenon is paralleled with the effect in MSK1-knockdown PC3 cells. In particular, MSK1 has been identified as the target gene of miR-148a, suggesting miR-148a attenuates the drug-resistance of CRPC cells via targeting MSK1 (135). Apparently, miR-148a represents a unique miRNA with dual function in PCa. Although the exact mechanism remains undetermined, AR may be an important factor involved in miR-148a function or the presence of different target genes of miRNA-184a in PCa cells derived from different origins.
miR-106b/miR-25
A study (136) using computational approach to identify PTEN-target miRNAs has identified miR-106b/miR-25 as a candidate that is concomitantly overexpressed with its host gene, minichromosome maintenance protein 7 (MCM7), which result in enhanced cell transformation and initiated prostatic intraepithelial neoplasia (PIN) progression of PCa. This study has a significant finding to show a cooperative expression of an oncomir cluster with its oncogenic host gene, which could simultaneously generate “two-hits” effect on the malignant transformation of normal cells. In addition, a study using LNCaP cell line has demonstrated that miR-106b/miR-25 cluster is associated with PCa progression by targeting caspase 7, apoptosis-related cysteine peptidase (CASP7) mRNA, which is down-regulated in both primary PCa and metastatic lesions (134). Overall, miR-106b/miR-25 cluster is an oncomir cluster and it is often found altered in its expression level between PIN, primary, and metastatic PCa (86,137).
Conclusions
miRNA represents a new mechanism of regulating gene expression at either post-transcriptional or translational levels. Aberrant alteration of miRNAs has been clearly demonstrated in PCa. However, knowing complex regulatory mechanism and relationship of miRNAs and their multiple target genes have further complicated their functionality during carcinogenesis. Therefore, carefully dissecting the mechanisms and functional role of each miRNA in heterogeneous PCa cells will certainly generate new information that could be applied as biomarker(s) and developed into novel therapeutic strategies.
Acknowledgements
Funding: This work was supported in part by grants from the United States Army (W81XWH-11-1-0491 to JTH).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29. [PubMed]
- Koivisto P, Kononen J, Palmberg C, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res 1997;57:314-9. [PubMed]
- Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, et al. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am J Pathol 1994;144:735-46. [PubMed]
- Henshall SM, Quinn DI, Lee CS, et al. Altered expression of androgen receptor in the malignant epithelium and adjacent stroma is associated with early relapse in prostate cancer. Cancer Res 2001;61:423-7. [PubMed]
- Watson PA, Chen YF, Balbas MD, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A 2010;107:16759-65. [PubMed]
- Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer 2011;18:R183-96. [PubMed]
- Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483-9. [PubMed]
- Bonaccorsi L, Muratori M, Carloni V, et al. Androgen receptor and prostate cancer invasion. Int J Androl 2003;26:21-5. [PubMed]
- Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J 2010;24:769-77. [PubMed]
- Wu JD, Haugk K, Woodke L, et al. Interaction of IGF signaling and the androgen receptor in prostate cancer progression. J Cell Biochem 2006;99:392-401. [PubMed]
- Léotoing L, Manin M, Monte D, et al. Crosstalk between androgen receptor and epidermal growth factor receptor-signalling pathways: a molecular switch for epithelial cell differentiation. J Mol endocrinol 2007;39:151-62. [PubMed]
- Culig Z, Hobisch A, Cronauer MV, et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res 1994;54:5474-8. [PubMed]
- Gregory CW, Johnson RT Jr, Presnell SC, et al. Androgen receptor regulation of G1 cyclin and cyclin-dependent kinase function in the CWR22 human prostate cancer xenograft. J Androl 2001;22:537-48. [PubMed]
- Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem 1997;272:29821-8. [PubMed]
- Voegel JJ, Heine MJ, Tini M, et al. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J 1998;17:507-19. [PubMed]
- Zhou G, Hashimoto Y, Kwak I, et al. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol Cell Biol 2003;23:7742-55. [PubMed]
- Chua SS, Ma Z, Ngan E, et al. Cdc25B as a steroid receptor coactivator. Vitam Horm 2004;68:231-56. [PubMed]
- Narayanan R, Jiang J, Gusev Y, et al. MicroRNAs are mediators of androgen action in prostate and muscle. PloS One 2010;5:e13637. [PubMed]
- Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol 2012;24:333-40. [PubMed]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A 2007;104:17719-24. [PubMed]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14:1902-10. [PubMed]
- Landais S, Landry S, Legault P, et al. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res 2007;67:5699-707. [PubMed]
- Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628-32. [PubMed]
- Fujita S, Ito T, Mizutani T, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 2008;378:492-504. [PubMed]
- Iliopoulos D, Jaeger SA, Hirsch HA, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010;39:493-506. [PubMed]
- Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 2009;23:2806-11. [PubMed]
- Wang Y, Hu X, Greshock J, et al. Genomic DNA copy-number alterations of the let-7 family in human cancers. PloS One 2012;7:e44399. [PubMed]
- Dong JT, Boyd JC, Frierson HF Jr. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate 2001;49:166-71. [PubMed]
- Suh SO, Chen Y, Zaman MS, et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 2011;32:772-8. [PubMed]
- Vrba L, Jensen TJ, Garbe JC, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PloS One 2010;5:e8697. [PubMed]
- Hulf T, Sibbritt T, Wiklund ED, et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene 2013;32:2891-9. [PubMed]
- Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008;7:2591-600. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009;11:228-34. [PubMed]
- Feng Y, Zhang X, Song Q, et al. Drosha processing controls the specificity and efficiency of global microRNA expression. Biochim Biophys Acta 2011;1809:700-7.
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 2005;19:2979-90. [PubMed]
- Kawamata T, Yoda M, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep 2011;12:944-9. [PubMed]
- Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437-41. [PubMed]
- Nadiminty N, Tummala R, Lou W, et al. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PloS One 2012;7:e32832. [PubMed]
- Kim HH, Kuwano Y, Srikantan S, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009;23:1743-8. [PubMed]
- Nadiminty N, Tummala R, Lou W, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem 2012;287:1527-37. [PubMed]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007;21:1025-30. [PubMed]
- Dong Q, Meng P, Wang T, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PloS One 2010;5:e10147. [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [PubMed]
- Kong D, Heath E, Chen W, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PloS One 2012;7:e33729. [PubMed]
- Xu B, Niu X, Zhang X, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 2011;350:207-13. [PubMed]
- Peng X, Guo W, Liu T, et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PloS One 2011;6:e20341. [PubMed]
- Clapé C, Fritz V, Henriquet C, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PloS One 2009;4:e7542. [PubMed]
- Huang S, Guo W, Tang Y, et al. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol Rep 2012;28:1831-7. [PubMed]
- Fuse M, Nohata N, Kojima S, et al. Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int J Oncol 2011;38:1093-101. [PubMed]
- Xu N, Papagiannakopoulos T, Pan G, et al. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009;137:647-58. [PubMed]
- Kong D, Li Y, Wang Z, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells 2009;27:1712-21. [PubMed]
- Slabáková E, Pernicová Z, Slavíčková E, et al. TGF-beta1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate 2011;71:1332-43. [PubMed]
- Liu YN, Yin JJ, Abou-Kheir W, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013;32:296-306. [PubMed]
- Viticchiè G, Lena AM, Latina A, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011;10:1121-31. [PubMed]
- Saini S, Majid S, Yamamura S, et al. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin Cancer Res 2011;17:5287-98. [PubMed]
- Boll K, Reiche K, Kasack K, et al. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 2013;32:277-85. [PubMed]
- Gandellini P, Folini M, Longoni N, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res 2009;69:2287-95. [PubMed]
- Verdoodt B, Neid M, Vogt M, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol 2013;43:307-14. [PubMed]
- Hagman Z, Haflidadottir BS, Ceder JA, et al. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br J Cancer 2013;108:1668-76. [PubMed]
- Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 2011;17:211-5. [PubMed]
- Kong D, Heath E, Chen W, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am J Transl Res 2012;4:14-23. [PubMed]
- Yamamura S, Saini S, Majid S, et al. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PloS One 2012;7:e29722. [PubMed]
- Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PloS One 2009;4:e6816. [PubMed]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008;105:13421-6. [PubMed]
- Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008;322:1695-9. [PubMed]
- Cao P, Deng Z, Wan M, et al. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer 2010;9:108. [PubMed]
- Tao J, Wu D, Xu B, et al. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep 2012;27:1967-75. [PubMed]
- Lin SL, Chiang A, Chang D, et al. Loss of mir-146a function in hormone-refractory prostate cancer. RNA 2008;14:417-24. [PubMed]
- Xu B, Wang N, Wang X, et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer. Prostate 2012;72:1171-8. [PubMed]
- Musumeci M, Coppola V, Addario A, et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011;30:4231-42. [PubMed]
- Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008;14:1271-7. [PubMed]
- Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009;28:1714-24. [PubMed]
- Noonan EJ, Place RF, Basak S, et al. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget 2010;1:349-58. [PubMed]
- Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle 2010;9:923-9. [PubMed]
- Reis ST, Pontes-Junior J, Antunes AA, et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol 2012;12:14. [PubMed]
- Li T, Li D, Sha J, et al. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun 2009;383:280-5. [PubMed]
- Shi XB, Xue L, Ma AH, et al. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 2011;71:538-49. [PubMed]
- Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A 2007;104:19983-8. [PubMed]
- Amir S, Ma AH, Shi XB, et al. Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PloS One 2013;8:e61064. [PubMed]
- Chen Y, Zaman MS, Deng G, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila) 2011;4:76-86. [PubMed]
- Lin D, Cui F, Bu Q, et al. The expression and clinical significance of GTP-binding RAS-like 3 (ARHI) and microRNA 221 and 222 in prostate cancer. J Int Med Res 2011;39:1870-5. [PubMed]
- Mercatelli N, Coppola V, Bonci D, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PloS One 2008;3:e4029. [PubMed]
- le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007;26:3699-708. [PubMed]
- Jalava SE, Urbanucci A, Latonen L, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012;31:4460-71. [PubMed]
- Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008;68:6162-70. [PubMed]
- Murata T, Takayama K, Katayama S, et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis 2010;13:356-61. [PubMed]
- Hudson RS, Yi M, Esposito D, et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene 2013;32:4139-47. [PubMed]
- Motoyama K, Inoue H, Nakamura Y, et al. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res 2008;14:2334-40. [PubMed]
- Thuault S, Tan EJ, Peinado H, et al. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 2008;283:33437-46. [PubMed]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008;14:1539-49. [PubMed]
- Nam Y, Chen C, Gregory RI, et al. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011;147:1080-91. [PubMed]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009;139:693-706. [PubMed]
- Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008;10:987-93. [PubMed]
- Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009;41:843-8. [PubMed]
- Boyerinas B, Park SM, Shomron N, et al. Identification of let-7-regulated oncofetal genes. Cancer Res 2008;68:2587-91. [PubMed]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 2005;25:9198-208. [PubMed]
- Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007;67:9762-70. [PubMed]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006;29:903-6. [PubMed]
- Chang TC, Zeitels LR, Hwang HW, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A 2009;106:3384-9. [PubMed]
- Kung HJ, Evans CP. Oncogenic activation of androgen receptor. Urol oncol 2009;27:48-52. [PubMed]
- Yamaguchi S, Yamahara K, Homma K, et al. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis 2011;219:468-74. [PubMed]
- Ahmed S, Nawshad A. Complexity in interpretation of embryonic epithelial-mesenchymal transition in response to transforming growth factor-beta signaling. Cells Tissues Organs 2007;185:131-45. [PubMed]
- Eastham AM, Spencer H, Soncin F, et al. Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res 2007;67:11254-62. [PubMed]
- Wei J, Xu G, Wu M, et al. Overexpression of vimentin contributes to prostate cancer invasion and metastasis via src regulation. Anticancer Res 2008;28:327-34. [PubMed]
- Luo Y, He DL, Ning L. Expression of “epithelial-mesenchymal transition” associated proteins in prostate cancer cell lines with different metastatic potentials and its significance. Zhonghua Nan Ke Xue 2006;12:696-700. [PubMed]
- Drake JM, Strohbehn G, Bair TB, et al. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell 2009;20:2207-17. [PubMed]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601. [PubMed]
- Gregory PA, Bracken CP, Smith E, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell 2011;22:1686-98. [PubMed]
- Xiong M, Jiang L, Zhou Y, et al. The miR-200 family regulates TGF-beta1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol 2012;302:F369-79. [PubMed]
- Moes M, Le Bechec A, Crespo I, et al. A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PloS One 2012;7:e35440. [PubMed]
- Qu Y, Li WC, Hellem MR, et al. MiR-182 and miR-203 induce mesenchymal to epithelial transition and self-sufficiency of growth signals via repressing SNAI2 in prostate cells. Int J Cancer 2013;133:544-55. [PubMed]
- Bhatnagar N, Li X, Padi SK, et al. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis 2010;1:e105. [PubMed]
- Tucci P, Agostini M, Grespi F, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A 2012;109:15312-7. [PubMed]
- Rokhlin OW, Scheinker VS, Taghiyev AF, et al. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther 2008;7:1288-96. [PubMed]
- Kobayashi Y, Absher DM, Gulzar ZG, et al. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res 2011;21:1017-27. [PubMed]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624-9. [PubMed]
- Hoffmann MJ, Engers R, Florl AR, et al. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther 2007;6:1403-12. [PubMed]
- Ren G, Baritaki S, Marathe H, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res 2012;72:3091-104. [PubMed]
- Sherwood ER, Van Dongen JL, Wood CG, et al. Epidermal growth factor receptor activation in androgen-independent but not androgen-stimulated growth of human prostatic carcinoma cells. Br J Cancer 1998;77:855-61. [PubMed]
- Shen J, Xia W, Khotskaya YB, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013;497:383-7. [PubMed]
- Man YG, Fu SW, Liu AJ, et al. Aberrant expression of chromogranin A, miR-146a, and miR-146b-5p in prostate structures with focally disrupted basal cell layers: an early sign of invasion and hormone-refractory cancer? Cancer genomics proteomics 2011;8:235-44. [PubMed]
- Xu B, Feng NH, Li PC, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate 2010;70:467-72. [PubMed]
- Hyytinen ER, Frierson HF Jr, Boyd JC, et al. Three distinct regions of allelic loss at 13q14, 13q21-22, and 13q33 in prostate cancer. Genes chromosomes cancer 1999;25:108-14. [PubMed]
- Tuxhorn JA, Ayala GE, Smith MJ, et al. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res 2002;8:2912-23. [PubMed]
- Chung LW, Chang SM, Bell C, et al. Co-inoculation of tumorigenic rat prostate mesenchymal cells with non-tumorigenic epithelial cells results in the development of carcinosarcoma in syngeneic and athymic animals. Int J Cancer 1989;43:1179-87. [PubMed]
- Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res 2009;69:7165-9. [PubMed]
- Kajanne R, Miettinen P, Tenhunen M, et al. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol 2009;35:1175-82. [PubMed]
- Abdulghani J, Gu L, Dagvadorj A, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol 2008;172:1717-28. [PubMed]
- Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 2007;282:23716-24. [PubMed]
- Badgwell DB, Lu Z, Le K, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene 2012;31:68-79. [PubMed]
- Li Y, Liu M, Zhang Y, et al. Effects of ARHI on breast cancer cell biological behavior regulated by microRNA-221. Tumour Biol 2013;34:3545-54. [PubMed]
- Coppola V, Musumeci M, Patrizii M, et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 2013;32:1843-53. [PubMed]
- Walter BA, Valera VA, Pinto PA, et al. Comprehensive microRNA profiling of prostate cancer. J Cancer 2013;4:350-7. [PubMed]
- Fujita Y, Kojima K, Ohhashi R, et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem 2010;285:19076-84. [PubMed]
- Poliseno L, Salmena L, Riccardi L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 2010;3:ra29. [PubMed]
- Szczyrba J, Loprich E, Wach S, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res 2010;8:529-38. [PubMed]


