Androgen receptor epigenetics
Introduction
The androgen receptor (AR) is a steroid receptor member of the larger nuclear receptor family. It is comprised of a large N-terminal domain (NTD) that can strongly stimulate transcription, a C-terminal ligand binding domain (LBD) that has a weaker transactivation function, a central DNA binding domain (DBD), and a short hinge region between the DBD and LBD that mediates functions including nuclear translocation. In the absence of androgen, the AR associates with an HSP90 chaperone complex in the cytoplasm. Similarly to other steroid hormone receptors, in response to androgen binding the AR LBD undergoes a conformational change that repositions helix 12 to generate a binding site for coactivator proteins that contain LXXLL-motifs (although this coactivator binding site in the AR LBD binds initially to an LXXLL-like peptide in the AR NTD). The liganded AR then forms a homodimer in the nucleus and binds to regulatory regions of multiple genes that are critical for prostate differentiation and for its normal function. Significantly, the consistent expression of AR in prostate cancer (PCa), and its continued activity in PCas that relapse after androgen deprivation therapy (castration-resistant prostate cancer, CRPC), indicate that at least a subset of these genes are also critical for PCa development and progression. However, the identity of the AR regulated genes that are critical for PCa remain unclear, and the extent to which AR acquires new functions during PCa development and progression remains to be determined.
AR binding to DNA is mediated by its DNA binding domain through recognition of a particular DNA sequence, referred to as an androgen responsive element (ARE). The AR then recruits multiple additional proteins that can modify chromatin structure and ultimately recruit and activate RNA polymerase II (Figure 1). Although this simple model is essentially correct, studies over the past several years have revealed that each step is regulated by multiple mechanisms involving large numbers of proteins. In particular, the central role of chromatin structure and histone posttranslational modifications in regulating the functions of AR and all other transcription factors is being elucidated. While the role of DNA methylation in the epigenetic regulation of gene expression is well established, it is now appreciated that changes in histone structure play a central and dynamic role in the epigenetic regulation of gene expression. The focus of this review is on the role of histones in the regulation of AR function in PCa development and progression.
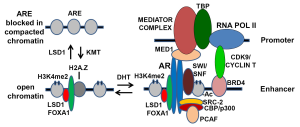
Through chromatin immunoprecipitation (ChIP) studies, coupled with massively parallel DNA sequencing (ChIP-seq) methods, it has been found that AR binds to thousands of sites in PCa cells and stimulates the expression of hundreds of genes. The first section below focuses on the role of chromatin structure in regulating AR binding to DNA. The second section then outlines how AR stimulates gene expression, with a focus on how it modulates chromatin structure. The third section describes the role of chromatin modifications in AR function as a transcriptional repressor. The subsequent sections then focus on epigenetic mechanisms that may alter the spectrum of AR regulated genes during PCa development and progression. The final section discusses possible therapeutic implications of AR epigenetics.
Epigenetic regulation of AR binding to chromatin
The AR DNA binding domain can directly mediate AR binding to AREs on naked DNA, but these sites are not readily available in vivo on compacted chromatin, which is tightly wound around nucleosomes. In order for most sites to become available for AR binding, the chromatin must first be “opened”, and this is generally achieved by binding of pioneer factors (1). Studies of steroid hormone receptors including ER in breast cancer and AR in PCa have established the central role of FOXA1 as a pioneer factor that binds initially to compacted chromatin and opens it for subsequent transcription factors (2-5). This capability of FOXA proteins is due to their structural similarity to linker histones, which allows them to bind between nucleosomes in compacted chromatin and locally open the chromatin (6,7). ChIP-seq studies in PCa cells have established that FOXA1 is associated with AR at most AR binding sites, and that FOXA1 is present at these sites prior to treatment with androgen to stimulate AR binding. Further studies have shown that these sites are DNAse hypersensitive, which is indicative of a nucleosome free region, prior to androgen treatment and AR binding (8,9).
Comparable results have been obtained for ER in breast cancer cells, and FOXA1 silencing by RNAi in breast cancer cells globally suppresses ER binding and activity (10). Interestingly, while FOXA1 silencing similarly impairs AR binding to a large fraction of sites in PCa cells, many sites are not affected and this also results in AR binding to new sites (11,12). The physiological significance of this observation remains uncertain, but it indicates that FOXA1 is not absolutely required for AR binding at all sites. It is possible that some of these sites may be pioneered by FOXA2, which is expressed during prostate development (13). AR binding sites are also highly enriched for the GATA2 and OCT1 transcription factors, and GATA2 may have a pioneering function on a subset of genes (4,7,9). Interestingly, FOXA1 mutations have been identified in a subset of advanced PCa (14,15). Therefore, modulation or alteration in FOXA1 is a possible mechanism that could contribute to altering AR function in advanced PCa (see below).
In addition to FOXA1, AR binding sites are enriched for nucleosomes in which lysine 4 on histone 3 has been mono- or dimethylated (H3K4me1 or H3K4me2) (3,8,16). A large body of literature has now established that H3K4me2 (and to a lesser extent H3K4me1) is specifically associated with transcriptional enhancers, while H3K4 trimethylation (H3K4me3) is associated with active promoters. Similarly to FOXA1, nucleosomes with the H3K4me2 mark are present at AR binding sites prior to androgen stimulation, and may contribute to the initial recruitment of FOXA1. However, the precise interplay between H3K4 methylation, FOXA1 binding, and likely other mechanisms in initiating the opening of an AR regulated enhancer remain to be clearly defined. Indeed, it is likely that this enhancer opening is not rigidly controlled, and that a balance between histone methylation, demethylation, and other modifications determines its status. In particular, amongst the many AR recruited proteins is a methyltransferase, SET9, that can methylate H3K4 and may thereby reinforce an open chromatin state (17,18). In contrast, H3K4me1 and H3K4me2 can be demethylated by the histone demethylase LSD1 (lysine specific demethylase 1, KDM1A), and it has been shown that LSD1 overexpression can inactivate AR regulated enhancers (19). Broader roles for LSD1 in AR functions as a transcriptional enhancer and repressor are described in subsequent sections below.
In contrast to H3K4 methylation that is generally associated with active enhancers (H3K4me2) and promoters (H3K4me3), methylation of H3K9 (H3K9me3) and H3K27 (H3K27me3) at promoters are strongly associated with transcriptional repression. Mechanistically, H3K9me3 can mediate interactions with proteins and long noncoding RNA (lncRNA) that localize the gene to transcriptionally inactive nuclear domains (20,21). AR has been found to interact with and stimulate the expression of KDM4B, an enzyme that can demethylate H3K9me3, and KDM4B can coactivate AR transcriptional activity (22). This coactivation may in part reflect H3K9me3 demethylation, but KDM4B also functions through a nonepigenetic mechanism to decrease the ubiquitylation and degradation of AR. In contrast to H3K9me3, H3K27me3 recruits DNA methyltransferases that methylate DNA, and can thereby mediate long-term gene silencing. H3K27 methylation is mediated by the Polycomb Repressive Complex 2 (PRC2), with EZH2 being the H3K27 methyltransferase in this complex. Previous studies established a strong correlation between increased EZH2 expression and more aggressive PCa (23,24). This correlation may reflect in part the progressive inactivation of AR regulated and AR independent genes that mediate differentiated functions and suppress growth. Indeed, mutations in another enzyme that can demethylate H3K27 (JMJD3, KDM6A) have been found in PCa, which may also contribute to gene inactivation (15). However, EZH2 was recently found to also function as an AR coactivator through a mechanisms that was methyltransferase dependent, but independent of PRC2 and H3K27 methylation (described further below) (25).
Epigenetic mechanisms through which AR stimulates gene expression
AR binds to enhancer sites that are generally distant from the promoter, but AR at these sites interacts with gene promoters by chromatin looping (26,27). A major contributor to this looping is the large multiprotein Mediator complex, a component of which interacts with AR (MED1) while other components interact with and include RNA polymerase II associated TATA binding proteins (28-31). Mutations in a Mediator protein (MED12) have been found in PCa, but the functional significance of these mutations is not clear (14). Additional proteins, as well as lncRNAs, likely contribute to the enhancer-promoter interaction. Interestingly, this looping mechanism has also been implicated in the generation of gene fusions occurring in PCa, including the TMPRSS2:ERG fusion (32,33).
Perhaps the earliest event that can be detected in response to AR binding is loss of a central nucleosome that overlaps the AR binding site, which can be demonstrated by ChIP and by an increase in DNase hypersensitivity, and is also associated with stronger binding of flanking nucleosomes (8,9). Interestingly, this central nucleosome located over the ARE at many AR stimulated enhancers contains the histone variant H2A.Z. This variant is also loaded onto nucleosomes during DNA repair and results in weaker nucleosome binding, which may facilitate the initial binding of AR. The androgen liganded AR then mediates the recruitment of multiple proteins that covalently modify histones and associated proteins, protein complexes that further unwind chromatin (primarily the SWI/SNF complex), proteins that mediate interaction with the promoter (see above), and ultimately enhance binding and activation of RNA polymerase II.
Many of the initially identified proteins recruited by AR, including the p160 steroid receptor coactivator proteins (SRC-1, 2 and 3), CBP and its close relative p300, and PCAF have lysine acetyltransferase activity and function as histone acetyltransferases (HATs), although it has become clear that they can also acetylate many additional proteins. Acetylation of lysines on histones may reduce their positive charge and thereby weaken their interaction with DNA. Acetylation at some sites may also prevent other modifications that repress gene expression, such as acetylation of H2K27 that would prevent EZH2 mediated trimethylation and gene silencing (see above). A further important function for histone lysine acetylation is the recruitment of proteins that contain bromodomains, which recognize acetyl-lysine residues (34). One such protein is BRD4, which contains two bromodomains and subsequently functions to recruit the CDK9/cyclin T complex (positive transcription elongation factor b, P-TEFb) that phosphorylates RNA polymerase II to drive elongation (35,36). Interestingly, CDK9 can also directly associate with and phosphorylate AR (37). Very recent studies indicate that BRD4 may function primarily on “super enhancers”, indicating that the AR-CDK9 interaction may play an important role in mediating interactions between AR bound to the enhancer and the promoter (chromatin looping, see above) for a subset of genes that are less dependent on BRD4.
Changes in histone acetylation (mediated by HATs and histone deacetylases, HDACs) occur rapidly and were initially viewed as the major posttranslational modification mediating the stimulation of transcription in response to hormone stimulation. In contrast, histone methylation on lysines was considered to function over a longer time frame and modulate enhancer availability. However, with the discovery of multiple enzymes that can demethylate histones, it now appears that androgen stimulated methylation of histone and nonhistone proteins also contributes acutely to gene activation. AR recruits and is coactivated by methyltransferases including the arginine methyltransferase CARM1 (PRMT4) (38) and the lysine methytransferase SET9 (17,18). CARM1 methylates arginine 17 on histone 3 (H3R17me2), but also has nonhistone substrates including SRC-3, and the precise basis for its AR coactivation function remains to be determined (39). As noted above, SET9 is recruited by AR to AREs and can methylate H3K4, and may thereby contribute to maintaining AR regulated enhancers. However, SET9 can also methylate lysines in the AR hinge region, which may enhance the interaction between the AR NTD and LBD. This direct effect on AR may account for SET9 coactivator function, but it will likely have additional substrates that remain to be defined.
As indicated in the previous section, LSD1 was identified initially as a demethylase for H3K4me1 and H3K4me2 (due to its catalytic mechanism, LSD1 can’t demethylate trimethylated lysines) (40), and was found to be associated with corepressor complexes (see below). However, it was subsequently found that LSD1 also functions as a critical coactivator for AR, as LSD1 inhibition or silencing by RNAi markedly decreased androgen stimulated expression of multiple AR regulated genes (41,42). Moreover, ChIP studies have demonstrated binding of LSD1 to AR regulated enhancers in these genes. Interestingly, while LSD1 can interact directly with AR, LSD1 binding to these enhancers (similarly to FOXA1 binding) is observed prior to androgen treatment, indicating that other interactions are mediating its recruitment. LSD1 may similarly be a coactivator for a subset of other transcription factors including ER (43,44).
One mechanism that may contribute to LSD1 function as a transcriptional coactivator is demethylation of repressive mono- and dimethylated H3K9 (41). Moreover, histone 3 phosphorylation may mediate a switch in the substrate specificity of LSD1 from H3K4 on AR repressed genes to H3K9 on AR stimulated genes. Specifically, phosphorylation of histone 3 on threonine 11 (H3T11ph) by protein kinase C-related kinase 1 (PRK1, PKN1) was found to enhance the ability of LSD1 to demethylate H3K9me1,2 (45). In contrast, phosphorylation of histone 3 on threonine 6 (H3T6ph) by protein kinase C 1 (PKC1) was found to inhibit LSD1 mediated demethylation of H3K4me1,2 (46). Significantly, both these kinases were found to associate with AR and be recruited by AR to androgen stimulated enhancers, supporting the model that they mediate a switch in LSD1 substrate specificity from H3K4 to H3K9. However, this switch is not absolute as LSD1 mediates some degree of H3K4 demethylation at androgen stimulated genes, and also mediates both H3K4 and H3K9 demethylation at AR repressed genes (see below) (47). Moreover, it is not clear whether alterations in H3K4 and H3K9 mono- or dimethylation at enhancer sites regulate transcription or are instead a consequence of transcriptional activity. In any case, it is likely that further mechanisms contribute to the coactivation function of LSD1 on androgen stimulated genes, which may include novel histone or nonhisotne substrates.
In addition to LSD1, AR has been reported to recruit the H3K9me3 demethylase JMJD2C (KDM4C, also termed GASC1) to the androgen stimulated PSA gene enhancer (42). Moreover, JHDM2A (KDM3A), an H3K9me1,2 demethylase, has been reported to be recruited to AR stimulated genes and enhance their transcription (48). Therefore, in addition to LSD1, additional histone demethylases may contribute to AR stimulated H3K9 demethylation and transcriptional activity.
Epigenetic mechanisms mediating AR repression of gene expression
In addition to its well established function as a transcriptional activator, AR can also decrease the expression of multiple genes. For a subset of genes this decrease in expression is due to AR binding and interference with other transcription factors including SP1, RUNX2, JUN, and SMAD3 (49-51), or with β-catenin (52-57). The agonist liganded AR also may function more directly as a transcriptional repressor through an epigenetic mechanism by recruiting corepressors that mediate histone deacetylation, including ALIEN, DAX1, HEY, AES, PHB, and SHP, although the role of these corepressors in modulating specific AR regulated genes remains to be determined (58-63).
In contrast to the ligand-dependent DNA binding by steroid receptors, DNA binding by the larger family of nonsteroidal nuclear receptors is ligand-independent and these nuclear receptors generally function as transcriptional repressors in the absence of ligand. This repression is mediated by the corepressors NCoR and SMRT, which are similarly associated with histone deacetylases. NCoR and SNRT contain extended LXXLL-like motifs (CoRNR boxes) that can bind to the unliganded coactivator binding site in the LBD of nuclear receptors, but are displaced after ligand binding. In contrast to other nuclear receptors and steroid receptors, the androgen liganded AR can also associate with NCoR and SMRT (64-67). This interaction is probably through a distinct site on the AR N-terminal domain, and downregulation of NCoR and SMRT can enhance activity of the agonist liganded AR. Significantly, the altered structure of the AR LBD generated by some AR antagonists may enhance NCoR and SMRT binding and contribute to antagonist activity (66-70).
Androgen mediated transcriptional repression also has been linked to histone methylation via AR recruitment of EZH2 and an increase in the EZH2 catalyzed repressive H3K27me3 mark (71,72). Conversely, AR can function directly as a transcriptional repressor through an interaction with LSD1 and histone demethylation (47). As outlined above, LSD1 can function as an AR coactivator, but it has been most extensively characterized as a corepressor that demethylates mono- and dimethylated lysine 4 on histone 3 (40). Consistent with this corepressor function, LSD1 associates with AR on AREs in many AR repressed genes and mediates demethylation of H3K4me1 and H3K4me2 in response to androgen (47). Amongst these genes that are directly repressed by AR in association with LSD1 are the AR gene itself, and genes regulating androgen synthesis (AKR1C3 and HSD17B6), which provides a negative feedback loop to regulate AR signaling. Significantly, a large proportion of other AR repressed genes are in pathways mediating DNA synthesis, which may reflect a physiological function of AR to suppress cell growth and drive differentiation (47).
LSD1 associates tightly with CoREST (REST corepressor 1, RCOR1), which may both stabilize LSD1 and stimulate its H3K4 demethylase activity (73). LSD1 and CoREST, in addition to proteins including HDAC1 and HDAC2, are components of the BHC corepressor complex that is recruited by the transcription factor REST to repress expression of neuronal genes in non-neuronal cells (74). LSD1 also is a component of another multiprotein corepressor complex, the NuRD complex, which similarly contains HDAC1 and HDAC2 (75,76). The JmjC family histone demethylase JARID1B (KDM5B), which can demethylate H3K4me3 (associated with promoters) and thereby generate the H3K4me2 substrate for LSD1, also has been identified as a component of the NuRD complex (77). LSD1 also may associate with additional protein or protein complexes, such as SIRT1 that mediates H4K16 deacetylation, or with lncRNA, to coordinately modify chromatin structure and repress gene expression (78,79). Therefore, AR transcriptional repression that is linked to LSD1 may be driven by additional epigenetic mechanisms mediated by multiple LSD1 associated proteins.
Epigenetic reprogramming of AR during PCa development
Frequent fusions between the strongly AR regulated TMPRSS2 gene and the Ets family transcription factor ERG gene, as well as additional fusions involving TMPRSS2 or other AR regulated genes, have established a genetic mechanism through which AR acquires new functions during PCa development (80). Several genes that may be directly regulated by ERG have been identified, but the precise mechanisms through which ERG drives PCa development have not been clear (81,82). Recent studies have identified epigenetic mechanisms through which ERG may drive PCa development. One reported downstream functions of ERG in PCa is to increase expression of EZH2, which may then mediate epigenetic gene silencing through H3K27 methylation (82). ERG was also reported to downregulate AR expression and transcriptional activity.
In contrast, studies in the TMPRSS2:ERG fusion positive VCaP human PCa model showed that ERG expression was increasing the number of genes that were stimulated by androgen (83). The most critical ERG dependent AR stimulated gene in VCaP cells was found to be the SOX9 transcription factor. SOX9 regulates ductal morphogenesis in fetal prostate and maintenance of stem/progenitor cells in adult tissues (84-87). In genetically engineering mouse models, SOX9 knockdown can impair PCa development driven by MYC and SV40 T antigen (84), while SOX9 overexpression in prostate on a Pten-/+ background results in high grade dysplastic lesions that can progress to invasive PCa (83,88). Mechanistically, ERG appears to be functioning as a pioneer factor by binding to a site 3' of the SOX9 gene, with subsequent binding of FOXA1 and opening of an adjacent cryptic AR binding site.
These findings suggested that the oncogenic effects of ERG in prostate specific ERG overexpression mouse models may be mediated through a similar mechanism. Indeed, a recent study showed that ERG expression in mouse prostate, similarly to ERG in human PCa cells, reprograms AR to stimulate the expression of multiple new genes (89). However, SOX9 mRNA is not increased by ERG overexpression in mouse prostate, which may account for the weaker phenotype of transgenic ERG versus SOX9 overexpression in mouse prostate. Consistent with this finding, the ERG and AR binding site identified at the 3' end of the human SOX9 gene is not conserved in mouse (83). Interestingly, while ERG does not directly increase SOX9 expression, a recent study suggests that it may indirectly enhance SOX9 activity (90). In any case, these findings taken together indicate that epigenetic reprogramming of AR transcriptional activity contributes to PCa pathogenesis in at least a subset of cases.
Epigenetic reprogramming of AR in advanced CRPC
AR can also acquire new transcriptional activities by epigenetic mechanisms in advanced CRPC. AR in an LNCaP-derived CRPC cell line (LNCaP-abl) was found to stimulate the expression of multiple genes that were not AR regulated in the parental LNCaP cells. The novel AR transcriptional program in LNCaP-abl cells included multiple M-phase cell cycle genes such as CDK1 and UBE2C, which are also overexpressed in CRPC (19). Significantly, this was not just secondary to increased proliferation, as ChIP-seq studies showed that the androgen stimulated expression of these genes in LNCaP-abl cells was associated with increased AR binding to sites linked to these genes. There were also increased levels of H3K4me1,2 at AR binding sites in these genes, indicating that these AR regulated enhancers had been opened by an epigenetic mechanism. Consistent with this conclusion, overexpression of LSD1 could decrease H3K4 methylation and AR binding to these sites.
It is not yet clear whether a pioneer factor or other specific mechanisms are initiating a precise new AR transcriptional program in these cells, versus positive selection leading to the gradual outgrowth of cells that have activated genes mediating proliferation through a variety of mechanisms. However, a recent study uncovered a novel AR coactivator function for EZH2 in CRPC cells that may contribute to AR reprogramming (25). As noted above, EZH2 is a histone methyltransferase associated with the PRC2, and its increased expression is correlated with higher grades and more advanced PCa. EZH2 expression is similarly increased in other cancers, which may reflect progressive silencing of tumor suppressor genes through H3K27 trimethylation. However, this study showed that increased EZH2 in more advanced PCa was not associated with increased H3K27me3. Instead, EZH2 in CRPC cells was found to form a complex with AR that was recruited to genes including CDK1 and UBE2C. Moreover, it functioned as an AR coactivator by a methyltransferase dependent mechanism that was distinct from its ability to methylate H3K27 (25). This coactivator function of EZH2 was dependent on AKT mediated phosphorylation of serine 21 on EZH2. Phosphorylation of this site on EZH2 was shown previously to suppress its ability to methylate H3K27. It is presumed that this AR coactivator function of EZH2 is mediated through methylation of other substrates, which may include AR, but further studies are needed to identify these alternative substrates.
While the above AR reprogramming appears to be dependent on H3K4 methylation and FOXA1, the AR transcriptional program may also be altered by a distinct mechanism involving downregulation of FOXA1. Recent studies found that FOXA1 downregulation by RNAi caused the expected loss of many AR binding sites, but the unexpected result was a large number of new AR binding sites (11,12). Consistent with their FOXA1 independence, these new AR binding sites were not enriched for H3K4 methylation. However, they appear to be functional enhancers based on production of short enhancer-templated non-coding RNA (eRNA), and AR binding to a subset could stimulate enhancer-promoter looping and gene expression. Interestingly, motif analyses show that these new AR binding sites are enriched for strong consensus AREs, which may be important for FOXA1 independent AR binding. Together these findings indicate that FOXA1, while having an important role as a pioneer factor for AR binding to a large number of genes, may also function to suppress AR binding to another set of genes.
The clinical significance of these findings remains to be determined, but it is intriguing that FOXA1 mutations occur in a subset of PCa and could be driving tumor progression through this AR reprogramming mechanism. Mutations in enzymes mediating H3K4 methylation, MLL2 (KMT2D) and MLL3 (KMT2C) have also been found in PCa, and could possibly function in part by closing some H3K4me2/FOXA1 dependent enhancers and redirecting AR to FOXA1 independent sites (14,15,91). Finally, recent data indicate that AR splice variants lacking the LBD, which are increased in CRPC, may regulate a distinct set of genes that include genes driving cell cycle progression (92). These findings could reflect novel interactions between AR splice variants and EZH2 or FOXA1, but further studies are needed to determine their molecular basis (93,94).
Clinical implications of AR epigenetics
Current efforts to ablate AR focus on reducing androgen synthesis and developing direct AR antagonists. However, these approaches are not selective and instead broadly suppress AR stimulated regulated genes, many of which do not mediate tumor growth and some of which may suppress tumor growth. Moreover, these therapies may also enhance the expression of AR repressed genes. Data outlined in this review show that the spectrum of genes regulated by AR is not hard-wired, and that epigenetic modifications can have a profound effect on the genes AR stimulates or represses. Therefore, as an alternative approach, it may be possible to develop agents that can epigenetically modify the spectrum of genes the AR regulates, and possibly thereby enhance its differentiation functions. Such approaches could be of particular value for PCa prevention or for treatment of early disease. However, a better understanding of the epigenetic mechanisms regulating AR functions may be needed, and in particular it is not clear whether genes mediating specific functions or pathways are controlled by distinct epigenetic mechanisms. One possible approach to address this question may be to further characterize AR function in other tissues where AR clearly regulates distinct repertoires of genes, and determine the epigenetic basis AR functions in these tissues.
Acknowledgements
Funding: Work from the authors cited in this review has been supported by awards from the National Institutes of Health (R01 DK079962 to X.Y. and K99 CA166507 to C.C.), SPORE in Prostate Cancer P50 CA090381, Department of Defense Prostate Cancer Research Program, and the Prostate Cancer Foundation.
The authors apologize to the many colleagues whose work we were unable to discuss or cite.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer 2012;12:381-5. [PubMed]
- Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005;122:33-43. [PubMed]
- Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008;132:958-70. [PubMed]
- Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 2007;27:380-92. [PubMed]
- Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J 2011;30:3885-94. [PubMed]
- Clark KL, Halay ED, Lai E, et al. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993;364:412-20. [PubMed]
- Cirillo LA, Lin FR, Cuesta I, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 2002;9:279-89. [PubMed]
- He HH, Meyer CA, Shin H, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet 2010;42:343-7. [PubMed]
- Andreu-Vieyra C, Lai J, Berman BP, et al. Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol Cell Biol 2011;31:4648-62. [PubMed]
- Hurtado A, Holmes KA, Ross-Innes CS, et al. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43:27-33. [PubMed]
- Wang D, Garcia-Bassets I, Benner C, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011;474:390-4. [PubMed]
- Sahu B, Laakso M, Ovaska K, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J 2011;30:3962-76. [PubMed]
- Kaestner KH. The FoxA factors in organogenesis and differentiation. Curr Opin Genet Dev 2010;20:527-32. [PubMed]
- Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685-9. [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [PubMed]
- Lupien M, Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer 2009;16:381-9. [PubMed]
- Ko S, Ahn J, Song CS, et al. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol 2011;25:433-44. [PubMed]
- Gaughan L, Stockley J, Wang N, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 2011;39:1266-79. [PubMed]
- Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009;138:245-56. [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [PubMed]
- Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011;147:773-88. [PubMed]
- Coffey K, Rogerson L, Ryan-Munden C, et al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res 2013;41:4433-46. [PubMed]
- Yu J, Rhodes DR, Tomlins SA, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res 2007;67:10657-63. [PubMed]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624-9. [PubMed]
- Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012;338:1465-9. [PubMed]
- Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 2005;19:631-42. [PubMed]
- Wu D, Zhang C, Shen Y, et al. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol Metab 2011;22:474-80. [PubMed]
- Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010;467:430-5. [PubMed]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010;11:761-72. [PubMed]
- Wang Q, Sharma D, Ren Y, et al. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem 2002;277:42852-8. [PubMed]
- Chen Z, Zhang C, Wu D, et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J 2011;30:2405-19. [PubMed]
- Lin C, Yang L, Tanasa B, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 2009;139:1069-83. [PubMed]
- Mani RS, Tomlins SA, Callahan K, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science 2009;326:1230. [PubMed]
- Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012;12:465-77. [PubMed]
- Yang Z, Yik JH, Chen R, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005;19:535-45. [PubMed]
- Jang MK, Mochizuki K, Zhou M, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005;19:523-34. [PubMed]
- Gordon V, Bhadel S, Wunderlich W, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol 2010;24:2267-80. [PubMed]
- Teyssier C, Ou CY, Khetchoumian K, et al. Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Mol Endocrinol 2006;20:1276-86. [PubMed]
- Feng Q, Yi P, Wong J, et al. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 2006;26:7846-57. [PubMed]
- Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941-53. [PubMed]
- Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005;437:436-9. [PubMed]
- Wissmann M, Yin N, Muller JM, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 2007;9:347-53. [PubMed]
- Garcia-Bassets I, Kwon YS, Telese F, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007;128:505-18. [PubMed]
- Wang J, Scully K, Zhu X, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007;446:882-7. [PubMed]
- Metzger E, Yin N, Wissmann M, et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol 2008;10:53-60. [PubMed]
- Metzger E, Imhof A, Patel D, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature 2010;464:792-6. [PubMed]
- Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20:457-71. [PubMed]
- Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006;125:483-95. [PubMed]
- Curtin D, Jenkins S, Farmer N, et al. Androgen suppression of GnRH-stimulated rat LHbeta gene transcription occurs through Sp1 sites in the distal GnRH-responsive promoter region. Mol Endocrinol 2001;15:1906-17. [PubMed]
- Verras M, Lee J, Xue H, et al. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res 2007;67:967-75. [PubMed]
- Grosse A, Bartsch S, Baniahmad A. Androgen receptor-mediated gene repression. Mol Cell Endocrinol 2012;352:46-56. [PubMed]
- Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res 2000;60:4709-13. [PubMed]
- Shah S, Hecht A, Pestell R, et al. Trans-repression of beta-catenin activity by nuclear receptors. J Biol Chem 2003;278:48137-45. [PubMed]
- Yang F, Li X, Sharma M, et al. Linking beta-catenin to androgen-signaling pathway. J Biol Chem 2002;277:11336-44. [PubMed]
- Chesire DR, Isaacs WB. Ligand-dependent inhibition of beta-catenin/TCF signaling by androgen receptor. Oncogene 2002;21:8453-69. [PubMed]
- Mulholland DJ, Read JT, Rennie PS, et al. Functional localization and competition between the androgen receptor and T-cell factor for nuclear beta-catenin: a means for inhibition of the Tcf signaling axis. Oncogene 2003;22:5602-13. [PubMed]
- Chen SY, Wulf G, Zhou XZ, et al. Activation of beta-catenin signaling in prostate cancer by peptidyl-prolyl isomerase Pin1-mediated abrogation of the androgen receptor-beta-catenin interaction. Mol Cell Biol 2006;26:929-39. [PubMed]
- Yuan X, Lu ML, Li T, et al. SRY interacts with and negatively regulates androgen receptor transcriptional activity. J Biol Chem 2001;276:46647-54. [PubMed]
- Jouravel N, Sablin E, Arnold LA, et al. Interaction between the androgen receptor and a segment of its corepressor SHP. Acta Crystallogr D Biol Crystallogr 2007;63:1198-200. [PubMed]
- Moehren U, Papaioannou M, Reeb CA, et al. Alien interacts with the human androgen receptor and inhibits prostate cancer cell growth. Mol Endocrinol 2007;21:1039-48. [PubMed]
- Gamble SC, Chotai D, Odontiadis M, et al. Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene 2007;26:1757-68. [PubMed]
- Belandia B, Powell SM, Garcia-Pedrero JM, et al. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol Cell Biol 2005;25:1425-36. [PubMed]
- Yu X, Li P, Roeder RG, et al. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol Cell Biol 2001;21:4614-25. [PubMed]
- Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol 2006;20:1048-60. [PubMed]
- Cheng S, Brzostek S, Lee SR, et al. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol Endocrinol 2002;16:1492-501. [PubMed]
- Hodgson MC, Astapova I, Cheng S, et al. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem 2005;280:6511-9. [PubMed]
- Song LN, Coghlan M, Gelmann EP. Antiandrogen effects of mifepristone on coactivator and corepressor interactions with the androgen receptor. Mol Endocrinol 2004;18:70-85. [PubMed]
- Hodgson MC, Shen HC, Hollenberg AN, et al. Structural basis for nuclear receptor corepressor recruitment by antagonist-liganded androgen receptor. Mol Cancer Ther 2008;7:3187-94. [PubMed]
- Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell 2002;9:601-10. [PubMed]
- Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol 2004;18:2633-48. [PubMed]
- Zhao JC, Yu J, Runkle C, et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res 2012;22:322-31. [PubMed]
- Chng KR, Chang CW, Tan SK, et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J 2012;31:2810-23. [PubMed]
- Shi YJ, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005;19:857-64. [PubMed]
- Hakimi MA, Bochar DA, Chenoweth J, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A 2002;99:7420-5. [PubMed]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 2011;11:588-96. [PubMed]
- Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009;138:660-72. [PubMed]
- Li Q, Shi L, Gui B, et al. Binding of the JmjC Demethylase JARID1B to LSD1/NuRD Suppresses Angiogenesis and Metastasis in Breast Cancer Cells by Repressing Chemokine CCL14. Cancer Res 2011;71:6899-908. [PubMed]
- Mulligan P, Yang F, Di Stefano L, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 2011;42:689-99. [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689-93. [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [PubMed]
- Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008;10:177-88. [PubMed]
- Yu J, Mani RS, Cao Q, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010;17:443-54. [PubMed]
- Cai C, Wang H, He HH, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest 2013;123:1109-22. [PubMed]
- Huang Z, Hurley PJ, Simons BW, et al. Sox9 is required for prostate development and prostate cancer initiation. Oncotarget 2012;3:651-63. [PubMed]
- Schaeffer EM, Marchionni L, Huang Z, et al. Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene 2008;27:7180-91. [PubMed]
- Thomsen MK, Butler CM, Shen MM, et al. Sox9 is required for prostate development. Dev Biol 2008;316:302-11. [PubMed]
- Wang H, Leav I, Ibaragi S, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res 2008;68:1625-30. [PubMed]
- Thomsen MK, Ambroisine L, Wynn S, et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res 2010;70:979-87. [PubMed]
- Chen Y, Chi P, Rockowitz S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 2013;19:1023-9. [PubMed]
- Wang G, Lunardi A, Zhang J, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet 2013;45:739-46. [PubMed]
- Kumar A, White TA, MacKenzie AP, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A 2011;108:17087-92. [PubMed]
- Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012;72:3457-62. [PubMed]
- Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 2012;287:19736-49. [PubMed]
- Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483-9. [PubMed]


