Cancer stem cells in prostate cancer
Introduction
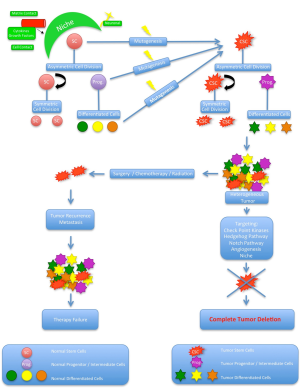
Despite enormous basic and clinical research efforts as well as progress in modern diagnosis and therapeutical options including surgery, radiation and chemotherapy the overall survival rates of human cancer barely increased in the last decades (1). Still patients die due to the continuous growth of metastases that most probably already occurred but were not detectable at the time of diagnosis. For a most effective cancer therapy it is critical to target the cancer cell sub-population with the ability for self-renewal, proliferation, invasive and metastatic growth. Many tumors contain phenotypically and functional heterogeneous cancer cells. In the traditional clonal evolution model tumors are believed to be homogenous and that all cells are able to repopulate and regenerate the tumor by themselves (2). Any heterogeneity is achieved by a subset of cells that acquire additional mutations after intrinsic (e.g., genetic, epigenetic) or extrinsic (e.g., microenvironment) stimuli that promote their aggressiveness and metastatic potential (3,4). As a consequence most of the currently used therapies aim to eliminate as many cancer cells as possible. Recently, a new model, arguing that tumors are malignant caricatures of normal development with an inherent hierarchical organization, presents an explanation for cancer heterogeneity (5). Only a small proportion of cells is capable of self-renewal and responsible for tumor initiation, growth and recurrence, while the majority of cells may be non-tumorigenic end cells. In parallel to normal tissues where cellular hierarchy is maintained by stem cells, this biologically distinct cancer cells have been termed cancer stem cells (CSCs, Figure 1). In this review we discuss the recent research in the field of CSCs, its limitations and therapeutical implications in general and specifically in P-Ca.
The history of CSC
The idea of a small subpopulation of cancer cells responsible for tumor initiation, hierarchical organization, growth and metastases has led to remarkable excitement in the field of cancer research, because it is thought to be responsible for the clinical observation that nearly all tumors are heterogeneous and that relapse often occurs in patients considered to be tumor free for many years. However, the idea has been around for some time. As early as 1855, Virchow postulated in his “embryonal rest hypothesis”, based on the histological similarities between teratocarcinomas and embryonic tissue, that the former originated from the latter (6). In 1937 Furth and Kahn described the transmission of leukemia in mice using only a single cell (7) and in the nineteen-sixties Pierce and colleagues demonstrated the clonal origin of mouse teratocarcinomas from single transplanted multipotent malignant cells (8). In 1997 Bonnet and Dick proved that human acute myeloid leukemias follow the CSC model (5). Single leukemia cell clones induced leukemia phenotypically identical to the parental tumor after transplanting them into immuno-deficient (NOD/SCID) mice. Those tumor-inducing cells revealed the surface markers CD34+/CD38–, characteristic of hematopoietic stem cells. Also solid tumor entities were described to follow the CSC hypothesis: Al-Hajj et al. transplanted only a few human breast cancer cells resulting in a tumor phenotypically identical to the original tumor. The tumor inducing cells (CD44+/CD24-/low) had in contrast to the non-tumor inducing cells (CD44–/CD24+) the capacity for self-renewal and massive proliferation (9). By today CSCs have been identified in many tumors including bladder (10), breast (9), brain (11), lung (12), prostate (13), ovary (14), colon (15), skin (16), liver (17), and other tumors (18).
CSC properties
Typically, CSCs are characterized to exhibit specific features similar to normal stem cells (SC): the ability for life-long and unlimited self-renewal allowing the maintenance of the CSC pool (19). This phenomenon is achieved by symmetric cell division into 2 new stem cells with the same fate. In contrast asymmetric cell division is thought to give rise to a new stem cell and a daughter cell that enters the differentiation process, loses multi-lineage potential and follows the hierarchical pattern (20). Experimentally, CSCs are defined by the ability to induce a phenotypic copy of the original tumor after serial transplantation into NOD/SCID mice (11). For some CSCs, self-preservation strategies including the activation of anti-apoptotic pathways, increased activity of membrane transporters, active drug efflux and enhanced DNA-repair activity has been described (21). Moreover the proposed ability of CSC to switch between an activated and quiescent state could serve as explanation for insufficient cancer therapies and long-term cancer recurrence (22), however the cell cycle distribution of most CSCs is unknown (23).
Identification/characterization of CSC
A common strategy for CSC identification is flow-cytometry using assumed specific CSC surface markers, e.g., CD44 or CD 133. However, many of the surface proteins used to identify CSCs are also expressed on physiological stem cells and/or progenitor cells (9,11,24). Moreover, since extensive research goes on discrepancies in marker expression of certain CSC entities as well as limited reproducibility has been reported, which could be due to differences in sample preparation and condition (fresh vs. passaged), dissociation techniques or even patient related (25,26). Marker based assays, especially single based, possibly enrich, but most probably do not isolate CSCs. Alternatively, different CSC clones with different marker expressions may coexist within primary tumors and/or functional different CSC clones might reside within defined cellular compartments (27,28). In order to reduce phenotypic variability the separation of live cancer cells based on functional measures e.g., signaling pathway activation has been demonstrated, however this is limited to mouse models with reduced variability using an inbred genetic background and targeted mutations (23). In label retaining assays all cells are labeled with a fluorescent marker which becomes more and more diluted with each cell division, therefore leaving the quiescent or low-cycling cell subpopulation positive (29). Utilizing the property of active efflux of the lipophilic dye (Hoechst 33342a) using ATP-binding cassette (ABC) transporters CSC containing “side populations” can be identified (30,31). In contrast to non-tumorigenic cells CSCs are able to form colonies from a single cell and have the ability to grow as spheres in serum free media (31,32). For genetic characterization of CSCs the expression of stemness genes as well as transcription factors can be used. Usually OCT4, Sox2 and Nanog are analyzed as they are essential for the maintenance of pluripotent embryonic stem cells (ESC). Other transcriptional factors are Bmi-1 (mediates gene silencing via regulation of chromatin structure) or Snail and Twist [promote epithelial-mesenchymal transition (EMT) (26,33,34)]. Xenograft models are considered to be the gold standard in the human CSC assay field. Mostly immuno-deficient mouse models are used due to the powerful xenogeneic immune response that kills most human cells before any proliferation. In these models CSCs are defined to have the ability to grow as serial transplantable tumors and to produce tumors showing the same biological heterogeneity as the parental tumor, hypothetically even after transplanting a single cell. However, these assays have limitations: the presence of species-specific signals, immune cells, tumor environment and niches as well as the site of injection are known to influence the efficiency of tumor initiation. In some cancers the transplantation into highly immune-deficient mice (NOD/SCID/IL-2Rγnull) can significantly increase the frequency of tumorigenic cells compared to transplantation into NOD/SCID mice, which retain an attenuated xenogeneic barrier (23,35). Therefore the different used mouse strains, method of tumor dissection and implantation influence experimental results and complicate the comparison of results.
CSC plasticity
The CSC concept should not be confused with the cell of origin. The cell of origin is the cell type first hit by an oncogenic mutation. Up to date the cell of origin for most cancers has not been yet precisely identified. CSCs can originate from stem cells through mutations that over-activate self-renewal mechanisms (36), however, it has also been demonstrated that they can arise from more differentiated progenitors. These acquire CSC properties by the accumulation of genetic and/or epigenetic abnormalities (37). Recent studies suggest that stemness may not be a fixed state but rather a flexible appearance (38). During the embryonic program of the epithelial-mesenchymal transition (EMT) cells acquire the ability to migrate, invade and to disseminate, which is mediated by transcriptional factors including Twist, Slug and Snail. The loss of epithelial markers and the gain of mesenchymal markers has been observed in epithelial cancer, whereas the overexpression of EMT regulators results in an enrichment of cells with CSC properties (39). In squamous cell carcinoma it has been demonstrated that CSC switch between a preferentially migratory or proliferative phenotype (40), leading to the theory of the existence of stationary and migratory CSCs and to the connection with circulating tumor cells (CTC). CTCs can be detected in blood from patients with primary and metastatic carcinomas. They are thought to be capable of self-seeding back to the original organs, which infers increased aggressiveness of the existing tumor or that they can settle in other organs such as bone marrow, a point at which they are termed disseminated tumor cells (DTC) and can serve as a reservoir of tumor cells responsible for future recurrence (41). Analogous to CSCs the EMT process is thought to be involved as the first step to allow the cells to enter circulation. The reverse process, [mesenchymal-epithelial transition (MET)] is thought to play a fundamental role after CTC have settled down in distant organs to form metastases in the new microenvironment (42). Both, CTCs and CSCs are able to become invasive, exhibit an increased level of resistance and stem cell like properties, enabling them to initiate metastatic growth (41). Whether CTCs and CSCs are entirely different populations is still a matter of debate; however EMT seems to be important and to link both entities. Another interesting possibility for the CSC origin comes from the discovery that the process of differentiation is reversible through the four transcription factors Klf4, Sox2, Oct4 and c-Myc. This so called Yamanaka-factors are highly expressed in ESC and their over-expression can induce pluripotency in both mouse and human somatic cells (iPSCs), giving differentiated cells the possibility to acquire (C)SC properties (43). Inspired by iPSC, so called induced cancer stem cells (iCSCs) from somatic cells have been established, that similar to iPSCs have the potential to undergo self-renewal, to generate differentiated progenies, and to form tumors when transplanted into recipient mice (44). The exact relations and shared mechanisms of CSCs, CTCs, ESCs and iPSCs are still unknown. However this data suggest that CSC most probably display a dynamic phenotype giving multiple options for the cell of origin.
CSC regulation
The CSC niche is a highly important regulatory anatomical microenvironment. Different cell types as well as a vascular network actively regulate CSC fate and plasticity e.g., by direct cell contact, matrix contact, extracellular matrix (ECM) components, cytokines and growth factors (45,46). For human medullblastoma cells it has been demonstrated that the CSC are located next to capillaries and the transplantation in xenografts with endothelial cells (EC) resulted in increased numbers of CSCs, tumor growth and production of vascular endothelial growth factor (VEGF) compared to transplantation without ECs (47). VEGF in turn leads to the production of EC, underscoring the bidirectional relationship of the nice and CSC. The ECM is essential for anchoring CSC to the niches and probably also modulate CSC function. There is an ongoing controversy whether the CSC can modify the composition of the ECM within the niche. Additionally the niche has a regulative role for CSC drug resistance, making it to an attractive target for new anti-cancer strategies (48,49). A number of different regulatory pathways and proteins orchestrate the fine balance of SC and CSC. From the nine main signaling pathways involved in embryonic development and cancer, seven of them have been implicated in both cancer and stem cells. These are: the JAK/STAT pathway, NOTCH signaling pathway, the MAP-Kinase/ERK pathway, the PI3K/AKT pathway, the NFkB pathway, the Wnt pathway and the TGFβ pathways (50). The dysregulation of signaling pathway networks plays an important role in enabling CSC to retain stem cell properties, however the detailed description is beyond the scope of this review.
Targeting CSC
Most of currently used anti-cancer therapies aim to kill cells in rapid expansion. Considering this from a CSC hypothesis point of view the results in a reduction of the non-tumorigenic tumor bulk, while the CSCs survive and later on may lead to metastasis and finally death. Treating leukemia in mice with valproic adic to induce growth arrest and apoptosis led to a fast tumor regression and prolonged animal survival, however after treatment withdrawal the disease recurred as a result of an increased self-renewal capacity of the CSCs (51). As mentioned above CSC self-preservation strategies include the activation of anti-apoptotic pathways, increased activity of membrane transporters, active drug efflux and enhanced DNA-repair activity. Since all these are potential targets multiple and different strategies are currently in the focus of extensive research (52). There is emerging evidence that CSCs enter quiescence in order to prevent self-renewal exhaustion. This would imply to either target the dormant CSC or to stimulate them to reenter the cell-cycle for a successful eradication. For chronic myeloid leukemia (CML) PML and FOXO were identified to be important regulators of quiescence. Targeting these genes in a mouse model resulted in an increase of CML proliferation and increased the sensitivity to the chemotherapeutic treatment with Imatinib (53). Therefore the combination of CSC stimulation and conventional drugs could be a reasonable treatment approach. Specifically aiming on CSC niches is an attractive concept as it simultaneously targets multiple signaling pathways, EMT and angiogenesis in the CSC microenvironment. In mice experiments the use of VEGF-Inhibitors led to a depletion of tumor blood vessels and reduction of medulloblastoma CSC (47). Clinically antiangiogenetic agents such as Bevacizumab improved outcomes in different cancer entities, especially in the combination with chemotherapy (54). Targeting surface markers has been demonstrated to successfully eradicate CSC, however it bears the risk of side effects as normal stem cells that exhibit the same markers are also targeted. A number of different regulatory pathways and proteins orchestrate the fine balance of SC and CSC. Out of many more the Wnt/b-catenin-, Notch-, PI3K/AKT/mTOR-, PTEN, and Hedgehog pathways are responsible for the maintenance of stemness, self-renewal, differentiation and resistance to treatment. Most of them are currently explored as targets for CSC eradication (55), for example the inhibition of notch signaling substantially reduced the CD 133+ brain CSC population in glioblastoma (56). However these pathways are highly conserved cell signaling systems and blocking them might have significant negative side effects. As mentioned above EMT is crucial for tumor metastasis. Experimentally EMT can be targeted with small RNAs. Micro RNAs are small non-coding RNAs that regulate the stability of mRNAs through interaction with the 3’ untranslated region of target genes (57). They are known to be important regulators for CSC self-renewal, differentiation, and tumorigenesis (58). In human breast CSC miR-200 inhibits TGF-b induced EMT, resulting in an inhibition of clonal expansion and tumor formation (59). Moreover one of the predicted target genes of miR-200c is Bmi-1, a regulator of self-renewal. In many cancers miR-34 expression is down regulated. Known targets of miR-34 are proteins important for apoptosis, cell cycle regulation and migration, mechanisms that are involved in CSC self-renewal and differentiation (60). Specifically one target gene is Bcl2, which improves CSC chemoresistance (61). Overexpressing miR-34 in pancreatic and gastric cancer sensitizes cells to chemotherapy, while tumor growth is inhibited (62,63). Moreover miR-34a leads to CSC differentiation through interaction with mRNA important for Notch1 and Notch2 in glioblastomas. Another currently unsolved problem is measuring CSC activity during and after treatment as specific markers/models still need to be established. However, they would be important for the correlation to outcome data in order to evaluate clinical efficacy.
The CSC hypothesis in P-Ca
Prostate stem cells (PSC)
The prostate consists of 3 cell types. Basal cells are relatively undifferentiated, androgen-independent, cells. They express CK 5 and 14, CD44 and p63, but no or only low androgen receptors (AR-), PSA and PAP. Secretory luminal, and glandular epithelial cells are differentiated cells of the mature prostate with androgen-receptor (AR+), PSA, PAP, CK 8 and 18 expression. The neuroendocrine cells appear to be androgen-independent and fully differentiated cells containing chromogranins without expression of AR or PSA (64). The existence of physiological stem cells in prostate was deduced from the finding that androgen ablation leads to involution of the androgen-dependent compartments of the prostate, but that subsequent androgen replacement results in total reconstitution of the organ. Based on these observations, Isaacs and Coffey developed a PSC model (65). According to this model, androgen-independent stem cells give rise to progenitor cells that are androgen-sensitive, but not androgen-dependent, which then, under the influence of androgen, differentiate into androgen-de- pendent cells of the prostate epithelium. Following Isaacs and Coffey’s theory the PSC were thought to reside in the basal cell layer, as it remains intact during androgen ablation-caused prostate involution.
Prostate CSC
CSCs in P-Ca are not well understood yet. There exists conflicting data for putative markers, the cell of origin as well as the location of P-Ca stem cells (PCSC) within the organ. However, ongoing research in mice and human provides convincing evidence that P-Ca follows the hierarchical model (66).
Markers used to study PSC and PCSC
Most studies investigating PCSC used established cell lines, primary tumors or xenografts in immuno-deficient mice (67). Multiple markers for the characterization of PSCs and PCSCs have been proposed, including cell surface markers, marker of self-renewal, pluripotency and markers of resistance to therapy (68). Collins isolated rare cells from human primary P-Ca using the combination of the surface markers CD44+α2β1integrinhighCD133+ that were able to self-renew in vitro (13). Using the same combination prostate CSCs were isolated from the cell line DU145 (69). Interestingly CD44, a glycoprotein involved in cell-cell interactions, cell adhesion and migration, has been identified as a marker of stemness of CSC for many different organs/cancer (70). Patrawala revealed that CD44+ P-Ca cells from xenograft human tumors were enriched in tumorigenic and metastatic progenitor cells compared to CD44- cells (71). Hurt demonstrated the tumor forming ability of CD44+CD24− prostate stem-like cells isolated from LNCaP cell line was after the injection of as few as 100 cells in NOD/SCID mice (72). Holoclones from the PC3 P-Ca cell line were shown to contain cells expressing high levels of CD44, α2β1 and β-catenin, and could initiate serially transplantable tumors after subcutaneous injection (73). CD133 has been identified as CSC marker for a variety of malignant tumors (74). In prostate, CD133+ cells were demonstrated to be able to possess a high in vitro proliferative potential and to reconstitute prostatic-like acini in immunocompromised male nude mice (24). However, recent studies suggest that CD133- cells in certain human tumors also possess tumorigenic activity after serial transplantation in NOD/SCID mice (74,75). Aldehyde dehydrogenase (ALDH1A1) acts in retinoic acid signaling, has important function in SC self-protection and high ALDH activity was correlated with the stem/progenitor cell state (76). For P-Ca it was found to be positively correlated with Gleason score and pathologic stage, and inversely associated with patient survival (77). In contrast to ALDH− cells, ALDH+ P-Ca cells showed CSC-like characteristics such as increased self-renewing and colony forming capacity and tumorigenicity. In addition, ALDH+ cells revealed an increased expression of putative P-Ca stem cell markers (CD44 and integrin α2β1) (78). Yu reported conflicting data, as they found that ALDHlowCD44- cells were also able to develop tumors with longer latency periods, although with lower capacity compared to their ALDHhighCD44+ counterparts (79). Investigating PSA−/loALDH+CD44+α2β1+ phenotypes Qin described these cells to be quiescent and refractory to stresses including androgen deprivation. The cells expressed stem cell genes, and were able to undergo asymmetric cell division generating PSA+ cells. Importantly they initiated robust tumor development, resisted androgen ablation and harbored highly tumorigenic castration-resistant PCa cells. In contrast, the PSA+ PCa cells possessed more limited tumor- propagating capacity, underwent symmetric division and were sensitive to castration (80). Lin-,Sca1+; CD49fhi (LSChi) cells have been demonstrated to be useful for isolation of murine stem cells. In the Pten-null P-Ca model the LSChi subpopulation is sufficient for cancer initiation (81). Addition of CD166 further enriched sphere-forming activity of WT LSChi and Pten null LSChi. Moreover expression of CD166 is upregulated in human P-Cas, especially CRPC samples (82). Nevertheless, in the Pten null mouse model downregulation of CD166 did interfere neither with sphere formation nor with progression and metastasis. Identifying the ABCG2 side population, which is associated with multidrug resistance, in combination with the surface marker CD133+/CD44+/CD24- have been also reported to increase CSC isolation (83). Another method to possibly identify the clinical relevant cell population is the exposure to chemotherapeutic agents such as Doxetacel. Using this method in DU145 and 22Rv1 cells with elevated levels of Notch and Hedgehog signaling were identified. Moreover these cells were also detected in human primary and metastatic prostate tumors (84). Taking another approach, an EMT phenotype with the loss of epithelial and gain of mesenchymal markers was described in isolated PC3 cells with CSC characteristics. Interestingly the cells overexpressed multiple stem cell genes such as Nanog, Oct4, and Sox2 (85). Recently Rajasekahr demonstrated the ability of a subpopulation of cells (TRA-1-60+CD151+CD166+) from human prostate xenografts to recapitulate the cellular hierarchy of the original tumor (86). The variability of the different marker combinations suggests that CSC may be more than a distinct subpopulation and underscores the idea of a dynamic CSC phenotype and plasticity.
Castration resistance
Androgen deprivation leads to reduction of the AR+ cell bulk of P-Ca (65,87). Castration resistant P-Ca expresses stem cell genes within the basal cell layer. The putative CD44+α2β1integrinhighCD133+ primary human prostate CSC identified by Collins et al. are AR-; they displayed a high capacity for self-renewal and differentiation into AR+ cells (13). The same was described for CD44+AR- tumor initiating cells from prostate xenografts that express stemness genes such as Oct3/4 or BMI1, suggesting a multi-lineage differentiation capacity (88,89). Lee described that P-Ca patients who received ADT had increased PCa stem/progenitor cells population. The addition of the anti-androgen, Casodex, or AR-siRNA in various PCa cells led to increased stem/progenitor cells, while in contrast, addition of functional AR led to decreased stem/progenitor cells population, but increased non-stem/progenitor cell population, suggesting that AR functions differentially in PCa stem/progenitor vs. non-stem/progenitor ßcells (90). This data propose that CSC in could contribute to castration resistance PCa. In the BM-18 xenograft model pre-existing stem cell (SC)-like and neuroendocrine (NE) PC cells are selected by castration and survive as totally quiescent and express the SC markers aldehyde dehydrogenase 1A1 (ALDH1A1) or NANOG, coexpress the luminal markers NKX3-1, CK18, and a low level of AR (ARlow), but not basal or NE markers (91).
The cell of origin
Stem cell biology and tumor biology are closely related, therefore lineage tracing studies in PSC can give insight in normal prostate regeneration, tumorigenesis and possibly the cell of origin in P-Ca, as this is highly relevant for understanding the applicability of a CSC model within the disease. According to the traditional model PSC reside in the basal layer, are AR- and give rise to AR+ luminal cells (92). Fitting into this model, intermediate cell types in transition between basal and luminal cells have been identified, expressing both basal and luminal markers (93). Stem cells in the basal layer are thought to be responsible for the regeneration of the prostate architecture after androgen ablation, however, this has also been demonstrated for a subset of luminal cells castration resistance (91,94). Recently, Zhou could show with the use of a mouse model for tracking cell fates and a mouse label-retaining assay that luminal cells are derived from a basal lineage and that slowly cycling cells, which may represent adult PSC, reside in the basal cell compartment (95). However for prostate CSC there is data supporting both, a basal and luminal cell of origin. In human PCa some believe that luminal cells are the cells of origin since the majority of cells in the tumor bulk are luminal and the disease is diagnosed based on the absence of basal markers. Moreover in human PIN the upregulation of c-MYC and shortening of telomere length was described exclusively in luminal but not in basal cells. Using a mouse Pten knockout P-Ca model all initial hyperplastic cells were luminal (67). Wang identified a rare luminal epithelial population with stem cell properties during prostate regeneration in mice, which they termed CARNs (Castration-resistant Nkx3.1-expressing cells). The deletion of Pten in CARNs resulted in high-grade PIN and carcinoma, indicating that CARNs are a cell of origin (96). The possibility of a human equivalent of CARNs was demonstrated by Germann et al. (91).
The CD44+α2β1integrinhighCD133+ CSC identified by Collins support the basal cell of origin theory as they comprised less than 1% of the tumor mass and were isolated from basal cells (13). Also the CD44+CD24− prostate stem-like cells described by Hurt revealed a basal phenotype (72). Cells within the basal fraction from human benign prostate tissues were able to regenerate benign prostate tissue in immuno-deficient mice. Interestingly the introduction of oncogenic alterations in the target cells induced a disease that mimics human P-Ca, while infected luminal cells failed to form tumors, supporting basal cells as one cell-of-origin for P-Ca (97). The fusion between the androgen receptor-regulated gene promoter of TMPRSS2 and ERG is present in about 50% of human P-Cas (98). In addition the TMPRSS2-ERG fusion was described to be present in the basal CD44+α2β1integrinhighCD133+ prostate CSC (99). Given the complexity and heterogeneity of prostate cancer it is likely that the different models (especially mouse models) only recapitulate properties of specific subtypes of human P-Ca (67). It has been speculated that there may also be multiple cells of origin for P-Ca in analogy with breast cancer (100). As this might lead to individual behavior and treatment response the investigation of cells of origin for P-Ca might have important clinical implications.
Targeting P-Ca stem cells
Similar to CSC in other cancer entities targeting of prostate CSC is subject of intensive research. As described above P-Ca patients who received ADT had increased PCa stem/progenitor cells population. Targeting PCa non-stem/progenitor cells with AR degradation enhancer ASC-J9® (GO-Y025, Dimethylcurcumin) and targeting PCa stem/progenitor cells with 5-azathioprine (immunosupressor) and gamma-tocotrieno (Vitamin E Isomer) resulted in significant suppression of the PCa at the castration resistant stage in human PCa cell lines and mouse models (90). This suggests a combinational therapy that simultaneously targets both stem/progenitor and non-stem/progenitor cells will lead to better efficacy. Targeting the hedgehog pathway Nanta investigated the effects of Erismodegib on human prostate CSC’s viability, sphere formation, apoptosis, EMT and tumor growth in NOD/SCID mice. The inhibition resulted in modulation of proliferation, tumor growth, EMT and apoptosis. Erismodegib inhibited CSC characteristics and regulation of Bcl-2 family members. Inhibition of Bmi-1 was meditated through upregulation of miR128 while the inhibition of EMT was regulated by induction of the miR-200 family. Targeting the hegdehog pathway could be a potential strategy for targeting prostate CSC (101).
Yang described a significantly higher expression of testicular nuclear receptor 4 (TR4) in PCa CD133+stem/progenitor cells compared with C133-non-stem/progenitor cells. The knockdown of TR4 led to increased drug sensitivity to two commonly used chemotherapeutic drugs, docetaxel and etoposide, mechanistic through the suppression of TR4 in these stem/progenitor cells led to down-regulation of Oct4 expression, which, in turn, downregulated the IL-1 receptor antagonist (IL1Ra) expression, suggesting the possibility of targeting TR4 as approach to overcome chemoresistance (102). MicroRNA profiling revealed that miR-34a is relatively lower expressed in CD44+ prostate CSCs from xenografts and primary tumors. The enforced expression of miR-34a in CD44+ cells inhibited clonogenic expansion, tumor regeneration and metastasis. In contrast miR-34a antagomirs in CD44- cells promoted tumor development and metastasis. Interestingly CD44 was described as a direct target of miR-34a. Therefore miR-34a could serve as therapeutic agent against prostate CSC (103).
Limitation of the CSC hypothesis
The CSC hypothesis is an attractive concept of cancer development and has led to some enthusiasm in the field of cancer research. It serves as logical explanation for clinical phenomenons such as tumor recurrence even years after an initially successful therapy. Most brilliant discoveries are simple, but now it appears that the more insight researcher gain into CSCs the more complex it gets. There are many theoretical and experimental caveats to the CSC model that have remained unexplored. For a detailed description we suggest the excellent review of Hans Clever and emphasize below the most important points (104). The above-mentioned plasticity of CSCs has yet not been understood in detail; however, the stability of the CSC phenotype is a precondition for selective targeting and plasticity might interfere with therapy. The species barrier as well as the transplantation setting limits the validity of the commonly used xenograft assays. Importantly, Morrison pointed out that the transplantation of any stem cell can reveal the potential of the stem cell under the particular assay conditions, but it cannot reveal the actual fate of the transplanted cell in its original tissue or tumor (23). The heterogeneity and inconsistency of the putative stem cell surface markers have already been discussed above. Often these heterogeneously expressed FACS markers were selected for their ability to isolate certain cells and not on the basis of a deeper understanding of the underlying stem cell biology of the pertinent tissue from which the cancer originates. Moreover it has been demonstrated that the tumorigenic cell frequencies can sometimes increase dramatically as a result of changes in assay conditions. Therefore it will be necessary to systematically assess the degree to which changes in assay conditions affect the spectrum of cancer cells that can form tumors.
Future perspectives
The exciting ongoing debate about the CSC theory will lead to further research elucidating the current controversies and open questions. Hopefully this will eventually result in the development of novel therapeutic strategies.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Howlader N, Noone AM, Krapcho M, et al. eds. SEER Cancer Statistics Review, 1975-2010. Available online: http://seer.cancer.gov/csr/1975_2010/
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [PubMed]
- Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 2011;17:320-9. [PubMed]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730-7. [PubMed]
- Virchow R. Cellular pathology. Arch Pathol Anat Physiol Klin Med 1855;8:3-39.
- Furth J, Kahn MC. The transmission of leukemia of mice with a single cell. Am J Cancer 1937;31:276-82.
- Kleinsmith LJ, Pierce GB Jr. Multipotentiality of single embryonal carcinoma cells. Cancer Res 1964;24:1544-51. [PubMed]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983-8. [PubMed]
- Volkmer JP, Sahoo D, Chin RK, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci U S A 2012;109:2078-83. [PubMed]
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396-401. [PubMed]
- Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005;121:823-35. [PubMed]
- Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-51. [PubMed]
- Bapat SA, Mali AM, Koppikar CB, et al. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res 2005;65:3025-9. [PubMed]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111-5. [PubMed]
- Fang D, Nguyen TK, Leishear K, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 2005;65:9328-37. [PubMed]
- Suetsugu A, Nagaki M, Aoki H, et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun 2006;351:820-4. [PubMed]
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol 2007;18:460-6. [PubMed]
- Lobo NA, Shimono Y, Qian D, et al. The biology of cancer stem cells. Annu Rev Cell Dev Biol 2007;23:675-99. [PubMed]
- Clevers H. Stem cells, asymmetric division and cancer. Nat Genet 2005;37:1027-8. [PubMed]
- Fábián A, Barok M, Vereb G, et al. Die hard: are cancer stem cells the Bruce Willises of tumor biology? Cytometry A 2009;75:67-74. [PubMed]
- Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol 2009;6:339-51. [PubMed]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 2012;21:283-96. [PubMed]
- Richardson GD, Robson CN, Lang SH, et al. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci 2004;117:3539-45. [PubMed]
- Nguyen LV, Vanner R, Dirks P, et al. Cancer stem cells: an evolving concept. Nat Rev Cancer 2012;12:133-43. [PubMed]
- Tirino V, Desiderio V, Paino F, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J 2013;27:13-24. [PubMed]
- Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086-93. [PubMed]
- Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol 2012;198:281-93. [PubMed]
- Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008;135:1118-29. [PubMed]
- Hirschmann-Jax C, Foster AE, Wulf GG, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A 2004;101:14228-33. [PubMed]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011;8:486-98. [PubMed]
- Rybak AP, He L, Kapoor A, et al. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim Biophys Acta 2011;1813:683-94.
- Young RA. Control of the embryonic stem cell state. Cell 2011;144:940-54. [PubMed]
- Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704-15. [PubMed]
- Quintana E, Shackleton M, Sabel MS, et al. Efficient tumour formation by single human melanoma cells. Nature 2008;456:593-8. [PubMed]
- Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8:511-24. [PubMed]
- Zhao Z, Zuber J, Diaz-Flores E, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 2010;24:1389-402. [PubMed]
- Iliopoulos D, Hirsch HA, Wang G, et al. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A 2011;108:1397-402. [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [PubMed]
- Biddle A, Liang X, Gammon L, et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res 2011;71:5317-26. [PubMed]
- Książkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: a hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology 2012;79:195-208. [PubMed]
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [PubMed]
- Nagata S, Hirano K, Kanemori M, et al. Self-renewal and pluripotency acquired through somatic reprogramming to human cancer stem cells. PLoS One 2012;7:e48699. [PubMed]
- Yang ZJ, Wechsler-Reya RJ. Hit ’em where they live: targeting the cancer stem cell niche. Cancer Cell 2007;11:3-5. [PubMed]
- Boral D, Nie D. Cancer stem cells and niche mircoenvironments. Front Biosci (Elite Ed) 2012;4:2502-14. [PubMed]
- Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007;11:69-82. [PubMed]
- Yi SY, Hao YB, Nan KJ, et al. Cancer stem cells niche: a target for novel cancer therapeutics. Cancer Treat Rev 2013;39:290-6. [PubMed]
- Kessenbrock K, Dijkgraaf GJ, Lawson DA, et al. A Role for Matrix Metalloproteinases in Regulating Mammary Stem Cell Function via the Wnt Signaling Pathway. Cell Stem Cell 2013;13:300-13. [PubMed]
- Dreesen O, Brivanlou AH. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev 2007;3:7-17. [PubMed]
- Leiva M, Moretti S, Soilihi H, et al. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia 2012;26:1630-7. [PubMed]
- Zhao L, Zhao Y, Bao Q, et al. Clinical implication of targeting of cancer stem cells. Eur Surg Res 2012;49:8-15. [PubMed]
- Zhang B, Strauss AC, Chu S, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell 2010;17:427-42. [PubMed]
- Bookman MA. The addition of new drugs to standard therapy in the first-line treatment of ovarian cancer. Ann Oncol 2010;21 Suppl 7:vii211-17. [PubMed]
- Alison MR, Lin WR, Lim SM, et al. Cancer stem cells: in the line of fire. Cancer Treat Rev 2012;38:589-98. [PubMed]
- Hovinga KE, Shimizu F, Wang R, et al. Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells 2010;28:1019-29. [PubMed]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 2006;103:4034-9. [PubMed]
- Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res 2011;71:5950-4. [PubMed]
- Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009;138:592-603. [PubMed]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010;17:193-9. [PubMed]
- Mathieu J, Ruohola-Baker H. Regulation of stem cell populations by microRNAs. Adv Exp Med Biol 2013;786:329-51. [PubMed]
- Ji Q, Hao X, Meng Y, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008;8:266. [PubMed]
- Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One 2009;4:e6816. [PubMed]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010;24:1967-2000. [PubMed]
- Isaacs JT, Coffey DS. Etiology and disease process of benign prostatic hyperplasia. Prostate Suppl 1989;2:33-50. [PubMed]
- Guzmán-Ramírez N, Völler M, Wetterwald A, et al. In vitro propagation and characterization of neoplastic stem/progenitor-like cells from human prostate cancer tissue. Prostate 2009;69:1683-93. [PubMed]
- Wang ZA, Shen MM. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene 2011;30:1261-71. [PubMed]
- Sharpe B, Beresford M, Bowen R, et al. Searching for prostate cancer stem cells: markers and methods. Stem Cell Rev 2013;9:721-30. [PubMed]
- Wei C, Guomin W, Yujun L, et al. Cancer stem-like cells in human prostate carcinoma cells DU145: the seeds of the cell line? Cancer Biol Ther 2007;6:763-8. [PubMed]
- Williams K, Motiani K, Giridhar PV, et al. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 2013;238:324-38. [PubMed]
- Patrawala L, Calhoun T, Schneider-Broussard R, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 2006;25:1696-708. [PubMed]
- Hurt EM, Kawasaki BT, Klarmann GJ, et al. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer 2008;98:756-65. [PubMed]
- Li H, Chen X, Calhoun-Davis T, et al. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res 2008;68:1820-5. [PubMed]
- Grosse-Gehling P, Fargeas CA, Dittfeld C, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol 2013;229:355-78. [PubMed]
- Stewart JM, Shaw PA, Gedye C, et al. Phenotypic heterogeneity and instability of human ovarian tumor-initiating cells. Proc Natl Acad Sci U S A 2011;108:6468-73. [PubMed]
- van den Hoogen C, van der Horst G, Cheung H, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res 2010;70:5163-73. [PubMed]
- Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest 2010;90:234-44. [PubMed]
- Yu C, Yao Z, Jiang Y, et al. Prostate cancer stem cell biology. Minerva Urol Nefrol 2012;64:19-33. [PubMed]
- Yu C, Yao Z, Dai J, et al. ALDH activity indicates increased tumorigenic cells, but not cancer stem cells, in prostate cancer cell lines. In Vivo 2011;25:69-76. [PubMed]
- Qin J, Liu X, Laffin B, et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell 2012;10:556-69. [PubMed]
- Mulholland DJ, Xin L, Morim A, et al. Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res 2009;69:8555-62. [PubMed]
- Jiao J, Hindoyan A, Wang S, et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 2012;7:e42564. [PubMed]
- Patrawala L, Calhoun T, Schneider-Broussard R, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 2005;65:6207-19. [PubMed]
- Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell 2012;22:373-88. [PubMed]
- Kong D, Banerjee S, Ahmad A, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One 2010;5:e12445. [PubMed]
- Rajasekhar VK, Studer L, Gerald W, et al. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun 2011;2:162. [PubMed]
- Tsujimura A, Koikawa Y, Salm S, et al. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J Cell Biol 2002;157:1257-65. [PubMed]
- Patrawala L, Calhoun-Davis T, Schneider-Broussard R, et al. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res 2007;67:6796-805. [PubMed]
- Gu G, Yuan J, Wills M, et al. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res 2007;67:4807-15. [PubMed]
- Lee SO, Ma Z, Yeh CR, et al. New therapy targeting differential androgen receptor signaling in prostate cancer stem/progenitor vs. non-stem/progenitor cells. J Mol Cell Biol 2013;5:14-26. [PubMed]
- Germann M, Wetterwald A, Guzmán-Ramirez N, et al. Stem-like cells with luminal progenitor phenotype survive castration in human prostate cancer. Stem Cells 2012;30:1076-86. [PubMed]
- Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev 2000;14:2410-34. [PubMed]
- van Leenders G, Dijkman H, Hulsbergen-van de Kaa C, et al. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab Invest 2000;80:1251-8. [PubMed]
- Wang G, Wang Z, Sarkar FH, et al. Targeting prostate cancer stem cells for cancer therapy. Discov Med 2012;13:135-42. [PubMed]
- Zhou J, Feigenbaum L, Yee C, et al. Mouse prostate epithelial luminal cells lineage originate in the basal layer where the primitive stem/early progenitor cells reside: implications for identifying prostate cancer stem cells. Biomed Res Int 2013;2013:913179.
- Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009;461:495-500. [PubMed]
- Goldstein AS, Huang J, Guo C, et al. Identification of a cell of origin for human prostate cancer. Science 2010;329:568-71. [PubMed]
- Mosquera JM, Mehra R, Regan MM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res 2009;15:4706-11. [PubMed]
- Birnie R, Bryce SD, Roome C, et al. Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol 2008;9:R83. [PubMed]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 2009;23:2563-77. [PubMed]
- Nanta R, Kumar D, Meeker D, et al. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rgamma null mice by regulating Bmi-1 and microRNA-128. Oncogenesis 2013;2:e42. [PubMed]
- Yang DR, Ding XF, Luo J, et al. Increased chemosensitivity via targeting testicular nuclear receptor 4 (TR4)-Oct4-interleukin 1 receptor antagonist (IL1Ra) axis in prostate cancer CD133+ stem/progenitor cells to battle prostate cancer. J Biol Chem 2013;288:16476-83. [PubMed]
- Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 2011;17:211-5. [PubMed]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011;17:313-9. [PubMed]